Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
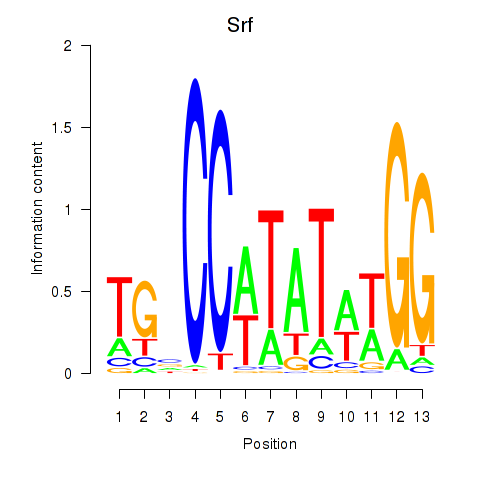
Results for Srf
Z-value: 0.61
Transcription factors associated with Srf
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Srf
|
ENSRNOG00000018232 | serum response factor |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Srf | rn6_v1_chr9_+_16737642_16737642 | 0.07 | 9.1e-01 | Click! |
Activity profile of Srf motif
Sorted Z-values of Srf motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_27657628 | 1.13 |
ENSRNOT00000026303
|
Egr1
|
early growth response 1 |
| chr20_-_22459025 | 0.57 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr4_-_117296082 | 0.45 |
ENSRNOT00000021097
|
Egr4
|
early growth response 4 |
| chr6_+_109300433 | 0.37 |
ENSRNOT00000010712
|
Fos
|
FBJ osteosarcoma oncogene |
| chr1_-_25839198 | 0.36 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr1_-_80221710 | 0.33 |
ENSRNOT00000091687
|
Fosb
|
FosB proto-oncogene, AP-1 transcription factor subunit |
| chr7_+_142912316 | 0.22 |
ENSRNOT00000010171
|
Nr4a1
|
nuclear receptor subfamily 4, group A, member 1 |
| chr1_-_72339395 | 0.21 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr1_-_47502952 | 0.18 |
ENSRNOT00000025580
|
Tagap
|
T-cell activation RhoGTPase activating protein |
| chr4_+_145330457 | 0.16 |
ENSRNOT00000011937
|
Arpc4
|
actin related protein 2/3 complex, subunit 4 |
| chr9_-_119332967 | 0.13 |
ENSRNOT00000021048
|
Myl12a
|
myosin light chain 12A |
| chr3_-_105512939 | 0.13 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr6_-_127248372 | 0.12 |
ENSRNOT00000085517
|
Asb2
|
ankyrin repeat and SOCS box-containing 2 |
| chr17_-_80577027 | 0.12 |
ENSRNOT00000057827
|
Rsu1
|
Ras suppressor protein 1 |
| chr9_+_84689150 | 0.12 |
ENSRNOT00000072242
|
Kcne4
|
potassium voltage-gated channel subfamily E regulatory subunit 4 |
| chr19_-_37916813 | 0.12 |
ENSRNOT00000026585
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr7_-_142260896 | 0.11 |
ENSRNOT00000073536
|
Smagp
|
small cell adhesion glycoprotein |
| chr3_+_119776925 | 0.11 |
ENSRNOT00000018549
|
Dusp2
|
dual specificity phosphatase 2 |
| chr7_+_12782491 | 0.10 |
ENSRNOT00000065093
|
Cnn2
|
calponin 2 |
| chr3_+_159569363 | 0.10 |
ENSRNOT00000064159
|
Tox2
|
TOX high mobility group box family member 2 |
| chrX_+_115351114 | 0.08 |
ENSRNOT00000047553
|
Hmg1l1
|
high-mobility group (nonhistone chromosomal) protein 1-like 1 |
| chr10_-_88060561 | 0.07 |
ENSRNOT00000019133
|
Krt19
|
keratin 19 |
| chr1_+_220826560 | 0.07 |
ENSRNOT00000027891
|
Fosl1
|
FOS like 1, AP-1 transcription factor subunit |
| chr2_-_195908037 | 0.07 |
ENSRNOT00000079776
|
Tuft1
|
tuftelin 1 |
| chr8_-_87282156 | 0.07 |
ENSRNOT00000087874
|
Filip1
|
filamin A interacting protein 1 |
| chr5_+_164808323 | 0.06 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr13_+_52662996 | 0.06 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr15_-_34693034 | 0.05 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr2_+_208750356 | 0.05 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr3_-_167759273 | 0.05 |
ENSRNOT00000049457
|
AABR07054721.1
|
|
| chr19_-_25345790 | 0.05 |
ENSRNOT00000010050
|
Zswim4
|
zinc finger, SWIM-type containing 4 |
| chr1_+_199664173 | 0.04 |
ENSRNOT00000054980
|
Tgfb1i1
|
transforming growth factor beta 1 induced transcript 1 |
| chr1_-_215838209 | 0.04 |
ENSRNOT00000050760
|
Igf2
|
insulin-like growth factor 2 |
| chr6_+_102048372 | 0.04 |
ENSRNOT00000013375
|
Eif2s1
|
eukaryotic translation initiation factor 2 subunit 1 alpha |
| chr2_+_189423559 | 0.04 |
ENSRNOT00000029076
|
Tpm3
|
tropomyosin 3 |
| chr2_+_208749996 | 0.04 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr19_-_11057254 | 0.04 |
ENSRNOT00000025559
|
Herpud1
|
homocysteine inducible ER protein with ubiquitin like domain 1 |
| chr9_+_81689802 | 0.04 |
ENSRNOT00000021432
|
Vil1
|
villin 1 |
| chr9_+_94228960 | 0.04 |
ENSRNOT00000083137
ENSRNOT00000080897 |
Akp3
|
alkaline phosphatase 3, intestine, not Mn requiring |
| chr13_-_36022197 | 0.04 |
ENSRNOT00000091280
|
Cfap221
|
cilia and flagella associated protein 221 |
| chr2_-_45077219 | 0.03 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr7_-_12707922 | 0.03 |
ENSRNOT00000013857
|
Misp
|
mitotic spindle positioning |
| chr3_-_147263275 | 0.03 |
ENSRNOT00000029620
|
Psmf1
|
proteasome inhibitor subunit 1 |
| chr3_+_1452644 | 0.03 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr15_-_2648551 | 0.03 |
ENSRNOT00000086864
|
Vdac2
|
voltage-dependent anion channel 2 |
| chr9_+_16737642 | 0.03 |
ENSRNOT00000061430
|
Srf
|
serum response factor |
| chr3_+_152857592 | 0.03 |
ENSRNOT00000027445
|
Myl9
|
myosin light chain 9 |
| chr8_+_117780891 | 0.03 |
ENSRNOT00000077236
|
Shisa5
|
shisa family member 5 |
| chr14_+_37918652 | 0.03 |
ENSRNOT00000088441
|
Tec
|
tec protein tyrosine kinase |
| chr1_-_31262466 | 0.03 |
ENSRNOT00000029531
|
Mrpl36
|
mitochondrial ribosomal protein L36 |
| chr1_-_151106802 | 0.03 |
ENSRNOT00000021971
|
Tyr
|
tyrosinase |
| chr15_+_56757315 | 0.03 |
ENSRNOT00000078013
|
Esd
|
esterase D |
| chr6_+_76745981 | 0.03 |
ENSRNOT00000076784
|
Brms1l
|
breast cancer metastasis-suppressor 1-like |
| chr10_-_13814304 | 0.03 |
ENSRNOT00000012203
|
Dnase1l2
|
deoxyribonuclease 1 like 2 |
| chr1_+_252497452 | 0.03 |
ENSRNOT00000075598
|
Stambpl1
|
STAM binding protein-like 1 |
| chr3_-_141411170 | 0.03 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr14_-_86146744 | 0.02 |
ENSRNOT00000019335
|
Myl7
|
myosin light chain 7 |
| chr10_+_10761477 | 0.02 |
ENSRNOT00000004176
|
Rogdi
|
rogdi homolog |
| chr19_-_15421404 | 0.02 |
ENSRNOT00000083684
|
Slc6a2
|
solute carrier family 6 member 2 |
| chr16_+_49266903 | 0.02 |
ENSRNOT00000014704
|
Slc25a4
|
solute carrier family 25 member 4 |
| chr3_+_80555196 | 0.02 |
ENSRNOT00000067318
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr8_+_60709851 | 0.02 |
ENSRNOT00000021817
|
Rcn2
|
reticulocalbin 2 |
| chr2_+_251534535 | 0.02 |
ENSRNOT00000080311
|
AABR07013701.1
|
|
| chr7_-_75288365 | 0.02 |
ENSRNOT00000036904
|
Ankrd46
|
ankyrin repeat domain 46 |
| chr8_+_49077053 | 0.02 |
ENSRNOT00000087922
|
Ift46
|
intraflagellar transport 46 |
| chr7_-_54778848 | 0.02 |
ENSRNOT00000005399
|
Glipr1
|
GLI pathogenesis-related 1 |
| chr10_+_53621375 | 0.02 |
ENSRNOT00000004147
|
Myh3
|
myosin heavy chain 3 |
| chr2_+_35977828 | 0.02 |
ENSRNOT00000076712
|
LOC108348103
|
serine protease inhibitor Kazal-type 5-like |
| chr4_+_55720010 | 0.01 |
ENSRNOT00000063987
|
Fscn3
|
fascin actin-bundling protein 3 |
| chr15_-_54906203 | 0.01 |
ENSRNOT00000020186
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr9_-_113846053 | 0.01 |
ENSRNOT00000017253
|
LOC100910483
|
ankyrin repeat domain-containing protein 12-like |
| chr8_+_48569328 | 0.01 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr1_-_175796040 | 0.01 |
ENSRNOT00000024060
|
Mrvi1
|
murine retrovirus integration site 1 homolog |
| chr9_+_119873170 | 0.01 |
ENSRNOT00000037587
|
Mettl4
|
methyltransferase like 4 |
| chr10_+_70262361 | 0.01 |
ENSRNOT00000064625
ENSRNOT00000076973 |
Unc45b
|
unc-45 myosin chaperone B |
| chrX_-_156440461 | 0.01 |
ENSRNOT00000083951
|
Rpl10
|
ribosomal protein L10 |
| chr2_+_60966789 | 0.01 |
ENSRNOT00000025490
|
Slc45a2
|
solute carrier family 45, member 2 |
| chr6_-_135112775 | 0.01 |
ENSRNOT00000086310
|
LOC103692716
|
heat shock protein HSP 90-alpha |
| chr1_-_193700673 | 0.01 |
ENSRNOT00000071314
|
Paip2l1
|
polyadenylate-binding protein-interacting protein 2-like 1 |
| chr4_-_99125111 | 0.01 |
ENSRNOT00000009184
|
Smyd1
|
SET and MYND domain containing 1 |
| chr6_-_102047758 | 0.01 |
ENSRNOT00000012101
|
Atp6v1d
|
ATPase H+ transporting V1 subunit D |
| chr4_+_136676254 | 0.00 |
ENSRNOT00000088070
|
Cntn6
|
contactin 6 |
| chrX_+_71342775 | 0.00 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Srf
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.1 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) regulation of progesterone biosynthetic process(GO:2000182) |
| 0.1 | 0.6 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 0.4 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.4 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0043435 | response to corticotropin-releasing hormone(GO:0043435) cellular response to corticotropin-releasing hormone stimulus(GO:0071376) |
| 0.0 | 0.1 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.0 | 0.1 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.1 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.0 | GO:0032057 | negative regulation of translational initiation in response to stress(GO:0032057) response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.1 | GO:0034372 | triglyceride-rich lipoprotein particle remodeling(GO:0034370) very-low-density lipoprotein particle remodeling(GO:0034372) |
| 0.0 | 0.0 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.0 | GO:1900222 | positive regulation of transcription by glucose(GO:0046016) negative regulation of beta-amyloid clearance(GO:1900222) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.1 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.0 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.6 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.4 | GO:0070412 | R-SMAD binding(GO:0070412) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.6 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.1 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |