Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
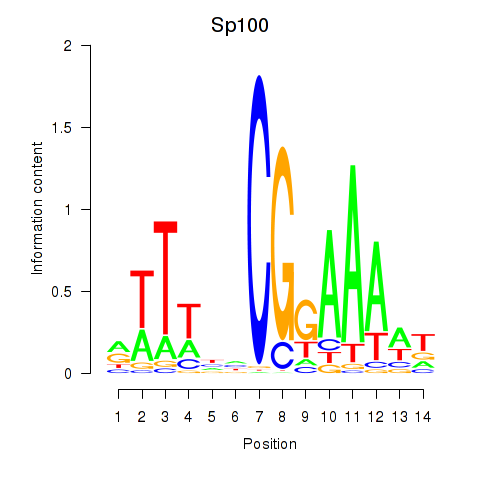
Results for Sp100
Z-value: 0.60
Transcription factors associated with Sp100
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sp100
|
ENSRNOG00000022769 | SP100 nuclear antigen |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sp100 | rn6_v1_chr9_+_92681078_92681078 | -0.71 | 1.8e-01 | Click! |
Activity profile of Sp100 motif
Sorted Z-values of Sp100 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_3793985 | 0.78 |
ENSRNOT00000049540
ENSRNOT00000086293 |
RT1-CE16
|
RT1 class I, locus CE16 |
| chr15_-_42794279 | 0.23 |
ENSRNOT00000023385
|
Ephx2
|
epoxide hydrolase 2 |
| chr2_+_206342066 | 0.23 |
ENSRNOT00000026556
|
Ptpn22
|
protein tyrosine phosphatase, non-receptor type 22 |
| chr2_-_113616766 | 0.21 |
ENSRNOT00000016858
ENSRNOT00000074723 |
Tmem212
|
transmembrane protein 212 |
| chr3_+_161121697 | 0.19 |
ENSRNOT00000036020
|
Wfdc10a
|
WAP four-disulfide core domain 10A |
| chr1_+_61786900 | 0.17 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr5_+_10125070 | 0.15 |
ENSRNOT00000085321
|
AABR07046866.1
|
|
| chr5_+_6373583 | 0.13 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr15_+_24159647 | 0.13 |
ENSRNOT00000082675
|
Lgals3
|
galectin 3 |
| chrM_+_3904 | 0.12 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chrX_+_25737292 | 0.12 |
ENSRNOT00000004893
|
RGD1563947
|
similar to RIKEN cDNA 4933400A11 |
| chr5_-_174921 | 0.12 |
ENSRNOT00000056239
|
Tipinl1
|
TIMELESS interacting protein like 1 |
| chr1_+_80056755 | 0.11 |
ENSRNOT00000021221
|
Snrpd2
|
small nuclear ribonucleoprotein D2 polypeptide |
| chr2_-_196415530 | 0.11 |
ENSRNOT00000064238
|
RGD1359334
|
similar to hypothetical protein FLJ20519 |
| chr12_-_38782010 | 0.11 |
ENSRNOT00000001813
|
Wdr66
|
WD repeat domain 66 |
| chr1_+_88620317 | 0.11 |
ENSRNOT00000075116
|
Cox7a1
|
cytochrome c oxidase subunit 7A1 |
| chr2_+_58724855 | 0.11 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr1_+_85496937 | 0.10 |
ENSRNOT00000076809
ENSRNOT00000087175 |
Selenov
|
selenoprotein V |
| chr4_-_23135354 | 0.10 |
ENSRNOT00000011432
|
Steap4
|
STEAP4 metalloreductase |
| chr1_+_66959610 | 0.10 |
ENSRNOT00000072122
ENSRNOT00000045994 |
Vom1r48
|
vomeronasal 1 receptor 48 |
| chrX_+_28593405 | 0.10 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chrM_+_9870 | 0.10 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr17_-_19703681 | 0.10 |
ENSRNOT00000024021
|
Mylip
|
myosin regulatory light chain interacting protein |
| chrM_+_11736 | 0.10 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr4_+_88834066 | 0.10 |
ENSRNOT00000009546
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr14_-_99780964 | 0.09 |
ENSRNOT00000006906
|
Sec61g
|
Sec61 translocon gamma subunit |
| chr1_-_62558033 | 0.09 |
ENSRNOT00000015520
|
Zfp40
|
zinc finger protein 40 |
| chr11_+_32440237 | 0.09 |
ENSRNOT00000040844
|
Kcne2
|
potassium voltage-gated channel subfamily E regulatory subunit 2 |
| chr1_-_247476827 | 0.09 |
ENSRNOT00000021298
|
Insl6
|
insulin-like 6 |
| chr4_-_59014416 | 0.09 |
ENSRNOT00000049811
|
RGD1561636
|
similar to 60S ribosomal protein L38 |
| chr10_-_14247886 | 0.09 |
ENSRNOT00000020504
|
Nubp2
|
nucleotide binding protein 2 |
| chr1_-_55895636 | 0.09 |
ENSRNOT00000029537
|
Vom2r9
|
vomeronasal 2 receptor, 9 |
| chr10_+_94566928 | 0.08 |
ENSRNOT00000078446
|
Prr29
|
proline rich 29 |
| chr11_-_32088002 | 0.08 |
ENSRNOT00000002732
|
Atp5o
|
ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit |
| chr2_-_205212681 | 0.08 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr6_+_93385457 | 0.08 |
ENSRNOT00000010300
|
Actr10
|
actin-related protein 10 homolog |
| chr12_-_20276121 | 0.08 |
ENSRNOT00000065873
|
LOC685157
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr9_+_51298426 | 0.08 |
ENSRNOT00000065212
|
Gulp1
|
GULP, engulfment adaptor PTB domain containing 1 |
| chr1_+_214016897 | 0.07 |
ENSRNOT00000075588
|
LOC100911766
|
tetraspanin-4-like |
| chr15_+_61879184 | 0.07 |
ENSRNOT00000042606
|
Sugt1
|
SGT1 homolog, MIS12 kinetochore complex assembly cochaperone |
| chr3_-_101547478 | 0.07 |
ENSRNOT00000006203
|
Fibin
|
fin bud initiation factor homolog (zebrafish) |
| chr8_-_85926450 | 0.07 |
ENSRNOT00000012474
|
Slc17a5
|
solute carrier family 17 member 5 |
| chr5_-_139933764 | 0.07 |
ENSRNOT00000015278
|
LOC680700
|
similar to ribosomal protein L10a |
| chr6_+_86131242 | 0.07 |
ENSRNOT00000039337
|
Fau
|
FAU, ubiquitin like and ribosomal protein S30 fusion |
| chr2_+_250600823 | 0.07 |
ENSRNOT00000083750
|
Sep15
|
selenoprotein 15 |
| chr10_-_83898527 | 0.07 |
ENSRNOT00000009815
|
Atp5g1
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr7_-_130350570 | 0.07 |
ENSRNOT00000055805
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr1_+_84652632 | 0.07 |
ENSRNOT00000041861
|
AABR07002774.2
|
|
| chr20_+_32450733 | 0.07 |
ENSRNOT00000036449
|
Rsph4a
|
radial spoke head 4 homolog A |
| chr20_+_34895059 | 0.07 |
ENSRNOT00000084420
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr13_-_73390393 | 0.07 |
ENSRNOT00000067253
ENSRNOT00000093438 |
Lhx4
|
LIM homeobox 4 |
| chr4_-_157750088 | 0.07 |
ENSRNOT00000038023
|
Cd27
|
CD27 molecule |
| chr4_+_25635765 | 0.07 |
ENSRNOT00000009340
|
Gtpbp10
|
GTP binding protein 10 |
| chr20_+_33945829 | 0.07 |
ENSRNOT00000064063
|
LOC103689994
|
radial spoke head protein 4 homolog A |
| chr2_-_30522727 | 0.06 |
ENSRNOT00000061183
|
Gtf2h2
|
general transcription factor IIH subunit 2 |
| chr1_+_224939497 | 0.06 |
ENSRNOT00000025664
|
Stx5
|
syntaxin 5 |
| chr19_+_14523554 | 0.06 |
ENSRNOT00000084271
ENSRNOT00000064731 |
Mcm5
|
minichromosome maintenance complex component 5 |
| chr5_-_92039146 | 0.06 |
ENSRNOT00000031852
|
Tmem261
|
transmembrane protein 261 |
| chr5_+_131257127 | 0.06 |
ENSRNOT00000010628
|
Bend5
|
BEN domain containing 5 |
| chr10_+_85257876 | 0.06 |
ENSRNOT00000014752
|
Mrpl45
|
mitochondrial ribosomal protein L45 |
| chrX_+_23668363 | 0.06 |
ENSRNOT00000004758
|
Gpr143
|
G protein-coupled receptor 143 |
| chr15_-_54906203 | 0.06 |
ENSRNOT00000020186
|
Cysltr2
|
cysteinyl leukotriene receptor 2 |
| chr9_+_69953440 | 0.06 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr8_+_58347736 | 0.06 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr15_+_57891680 | 0.06 |
ENSRNOT00000001383
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr5_-_118541928 | 0.06 |
ENSRNOT00000012947
|
Itgb3bp
|
integrin subunit beta 3 binding protein |
| chrM_+_7919 | 0.06 |
ENSRNOT00000046108
|
Mt-atp6
|
mitochondrially encoded ATP synthase 6 |
| chr6_+_108796182 | 0.06 |
ENSRNOT00000006297
|
Fcf1
|
FCF1 rRNA-processing protein |
| chr9_-_43251400 | 0.06 |
ENSRNOT00000037570
|
Fam178b
|
family with sequence similarity 178, member B |
| chr19_+_682065 | 0.06 |
ENSRNOT00000087469
|
Terb1
|
telomere repeat binding bouquet formation protein 1 |
| chr4_-_66624912 | 0.06 |
ENSRNOT00000064891
|
Hipk2
|
homeodomain interacting protein kinase 2 |
| chr1_-_222246765 | 0.06 |
ENSRNOT00000087285
ENSRNOT00000028731 |
Dnajc4
|
DnaJ heat shock protein family (Hsp40) member C4 |
| chr19_+_14345993 | 0.06 |
ENSRNOT00000084913
|
Gm18025
|
predicted gene, 18025 |
| chr12_-_21728678 | 0.06 |
ENSRNOT00000035372
|
LOC100910581
|
protein phosphatase 1 regulatory subunit 35-like |
| chr14_-_87701884 | 0.06 |
ENSRNOT00000079338
|
Mospd1
|
motile sperm domain containing 1 |
| chr9_+_17216495 | 0.06 |
ENSRNOT00000026331
|
Mad2l1bp
|
MAD2L1 binding protein |
| chr18_+_57654290 | 0.06 |
ENSRNOT00000025892
|
Htr4
|
5-hydroxytryptamine receptor 4 |
| chr10_-_38969501 | 0.06 |
ENSRNOT00000090691
ENSRNOT00000081309 ENSRNOT00000010029 |
Il4
|
interleukin 4 |
| chr1_-_221431713 | 0.06 |
ENSRNOT00000028485
|
Tm7sf2
|
transmembrane 7 superfamily member 2 |
| chr5_+_126668689 | 0.06 |
ENSRNOT00000036072
|
Cyb5rl
|
cytochrome b5 reductase-like |
| chr7_+_29909120 | 0.06 |
ENSRNOT00000049362
|
AABR07056556.1
|
|
| chr16_-_64745207 | 0.05 |
ENSRNOT00000032467
|
Tti2
|
TELO2 interacting protein 2 |
| chrX_-_135342996 | 0.05 |
ENSRNOT00000084848
ENSRNOT00000008503 |
Aifm1
|
apoptosis inducing factor, mitochondria associated 1 |
| chr4_+_100465170 | 0.05 |
ENSRNOT00000019571
|
Retsat
|
retinol saturase |
| chr15_-_34352673 | 0.05 |
ENSRNOT00000064916
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr4_-_81968832 | 0.05 |
ENSRNOT00000016608
|
Skap2
|
src kinase associated phosphoprotein 2 |
| chr17_+_43791657 | 0.05 |
ENSRNOT00000074024
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chr3_+_93351619 | 0.05 |
ENSRNOT00000011035
|
Elf5
|
E74-like factor 5 |
| chr13_+_83073544 | 0.05 |
ENSRNOT00000066119
ENSRNOT00000079796 ENSRNOT00000077070 |
Dpt
|
dermatopontin |
| chr4_-_100465106 | 0.05 |
ENSRNOT00000086725
ENSRNOT00000057989 |
Elmod3
|
ELMO domain containing 3 |
| chr9_-_82154266 | 0.05 |
ENSRNOT00000024283
|
Cryba2
|
crystallin, beta A2 |
| chr2_+_34312766 | 0.05 |
ENSRNOT00000060962
|
Cenpk
|
centromere protein K |
| chr1_-_167698263 | 0.05 |
ENSRNOT00000093046
|
Trim21
|
tripartite motif-containing 21 |
| chr18_+_86071662 | 0.05 |
ENSRNOT00000064901
|
Rttn
|
rotatin |
| chr1_-_162713610 | 0.05 |
ENSRNOT00000018091
|
Aqp11
|
aquaporin 11 |
| chr4_+_27698323 | 0.05 |
ENSRNOT00000012086
|
Rbm48
|
RNA binding motif protein 48 |
| chr10_-_70339578 | 0.05 |
ENSRNOT00000076618
|
Slfn13
|
schlafen family member 13 |
| chr1_-_62114672 | 0.05 |
ENSRNOT00000072607
|
AABR07001923.1
|
|
| chr1_+_274049533 | 0.05 |
ENSRNOT00000076779
ENSRNOT00000042822 |
Mxi1
|
MAX interactor 1, dimerization protein |
| chr5_+_135536413 | 0.05 |
ENSRNOT00000023132
|
Prdx1
|
peroxiredoxin 1 |
| chr10_-_104054805 | 0.05 |
ENSRNOT00000004853
|
Nt5c
|
5', 3'-nucleotidase, cytosolic |
| chr3_+_120414291 | 0.05 |
ENSRNOT00000083627
ENSRNOT00000021740 |
Acoxl
|
acyl-CoA oxidase-like |
| chr5_-_147828449 | 0.05 |
ENSRNOT00000083369
|
Ccdc28b
|
coiled coil domain containing 28B |
| chr6_+_29061618 | 0.05 |
ENSRNOT00000092223
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr2_-_90568486 | 0.05 |
ENSRNOT00000059380
|
LOC100910852
|
uncharacterized LOC100910852 |
| chr6_+_80108655 | 0.05 |
ENSRNOT00000006137
|
Gemin2
|
gem (nuclear organelle) associated protein 2 |
| chr6_-_111477090 | 0.05 |
ENSRNOT00000016389
|
Alkbh1
|
alkB homolog 1, histone H2A dioxygenase |
| chr6_+_72124417 | 0.05 |
ENSRNOT00000040548
|
Scfd1
|
sec1 family domain containing 1 |
| chr14_-_84662143 | 0.05 |
ENSRNOT00000057529
ENSRNOT00000080078 |
Hormad2
|
HORMA domain containing 2 |
| chr2_+_188583664 | 0.05 |
ENSRNOT00000045477
|
Dpm3
|
dolichyl-phosphate mannosyltransferase subunit 3 |
| chr20_-_4935372 | 0.04 |
ENSRNOT00000050099
ENSRNOT00000047779 |
RT1-CE3
RT1-CE4
|
RT1 class I, locus CE3 RT1 class I, locus CE4 |
| chr10_-_16689321 | 0.04 |
ENSRNOT00000028173
|
Bnip1
|
BCL2/adenovirus E1B interacting protein 1 |
| chr1_-_276012351 | 0.04 |
ENSRNOT00000045642
|
AABR07007000.1
|
|
| chr4_-_7529106 | 0.04 |
ENSRNOT00000013865
|
Nupl2
|
nucleoporin like 2 |
| chr15_-_52344629 | 0.04 |
ENSRNOT00000032832
|
Npm2
|
nucleophosmin/nucleoplasmin 2 |
| chr19_-_54245855 | 0.04 |
ENSRNOT00000023855
|
Emc8
|
ER membrane protein complex subunit 8 |
| chr12_-_39882030 | 0.04 |
ENSRNOT00000078761
ENSRNOT00000048851 |
Ppp1cc
|
protein phosphatase 1 catalytic subunit gamma |
| chr18_+_16544508 | 0.04 |
ENSRNOT00000020601
|
Elp2
|
elongator acetyltransferase complex subunit 2 |
| chr2_-_188252551 | 0.04 |
ENSRNOT00000088327
|
Dap3
|
death associated protein 3 |
| chr5_+_78384444 | 0.04 |
ENSRNOT00000034978
|
LOC500475
|
similar to hypothetical protein 4933430I17 |
| chr5_-_138629026 | 0.04 |
ENSRNOT00000049535
|
Rps27a
|
ribosomal protein S27a |
| chr8_-_77992621 | 0.04 |
ENSRNOT00000085843
|
Myzap
|
myocardial zonula adherens protein |
| chr11_+_38727048 | 0.04 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chrM_+_7758 | 0.04 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr19_-_914880 | 0.04 |
ENSRNOT00000017127
|
Cklf
|
chemokine-like factor |
| chr4_-_61720956 | 0.04 |
ENSRNOT00000012879
|
Akr1b1
|
aldo-keto reductase family 1 member B |
| chr12_+_22727335 | 0.04 |
ENSRNOT00000077293
|
Znhit1
|
zinc finger, HIT-type containing 1 |
| chr1_-_128695995 | 0.04 |
ENSRNOT00000077020
|
Synm
|
synemin |
| chr13_-_95943761 | 0.04 |
ENSRNOT00000005961
|
Adss
|
adenylosuccinate synthase |
| chrX_-_128268285 | 0.04 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr10_-_70259553 | 0.04 |
ENSRNOT00000011320
|
Nle1
|
notchless homolog 1 |
| chr2_-_3124543 | 0.04 |
ENSRNOT00000036547
|
Fam81b
|
family with sequence similarity 81, member B |
| chr1_-_80056574 | 0.04 |
ENSRNOT00000021200
|
Qpctl
|
glutaminyl-peptide cyclotransferase-like |
| chr12_+_30655898 | 0.04 |
ENSRNOT00000079144
|
Zfp11
|
zinc finger protein 11 |
| chr1_+_267618248 | 0.04 |
ENSRNOT00000017186
|
Gsto2
|
glutathione S-transferase omega 2 |
| chr1_-_91063928 | 0.04 |
ENSRNOT00000072756
|
Capns1
|
calpain, small subunit 1 |
| chr3_+_132560506 | 0.04 |
ENSRNOT00000005920
|
Sptlc3
|
serine palmitoyltransferase, long chain base subunit 3 |
| chr15_-_93667355 | 0.04 |
ENSRNOT00000093571
ENSRNOT00000089876 |
Fbxl3
|
F-box and leucine-rich repeat protein 3 |
| chr20_+_3556975 | 0.04 |
ENSRNOT00000089417
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr1_+_56982821 | 0.04 |
ENSRNOT00000059845
|
Ermard
|
ER membrane-associated RNA degradation |
| chr2_-_27364906 | 0.04 |
ENSRNOT00000078639
|
Polk
|
DNA polymerase kappa |
| chr8_-_2045817 | 0.04 |
ENSRNOT00000009542
ENSRNOT00000081171 ENSRNOT00000078765 |
Gria4
|
glutamate ionotropic receptor AMPA type subunit 4 |
| chr12_-_21989339 | 0.04 |
ENSRNOT00000074735
|
Ppp1r35
|
protein phosphatase 1, regulatory subunit 35 |
| chr20_+_5414448 | 0.04 |
ENSRNOT00000078972
ENSRNOT00000080900 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chr1_-_216750844 | 0.04 |
ENSRNOT00000054862
|
Nap1l4
|
nucleosome assembly protein 1-like 4 |
| chr20_+_6869767 | 0.04 |
ENSRNOT00000000631
ENSRNOT00000086330 ENSRNOT00000093188 ENSRNOT00000093736 |
RGD735065
|
similar to GI:13385412-like protein splice form I |
| chr16_-_70998575 | 0.04 |
ENSRNOT00000019935
|
Kcnu1
|
potassium calcium-activated channel subfamily U member 1 |
| chr4_-_71713063 | 0.04 |
ENSRNOT00000059447
|
Fam131b
|
family with sequence similarity 131, member B |
| chr2_-_185005572 | 0.04 |
ENSRNOT00000093291
|
Fam160a1
|
family with sequence similarity 160, member A1 |
| chr1_-_79816450 | 0.04 |
ENSRNOT00000018495
|
Ccdc61
|
coiled-coil domain containing 61 |
| chr5_+_128450680 | 0.04 |
ENSRNOT00000010700
|
Txndc12
|
thioredoxin domain containing 12 |
| chr10_-_98544447 | 0.04 |
ENSRNOT00000073149
|
Abca6
|
ATP binding cassette subfamily A member 6 |
| chr2_+_189609800 | 0.04 |
ENSRNOT00000089016
|
Slc39a1
|
solute carrier family 39 member 1 |
| chr3_-_82906581 | 0.04 |
ENSRNOT00000030784
|
Alkbh3
|
alkB homolog 3, alpha-ketoglutarate-dependent dioxygenase |
| chr12_-_22006533 | 0.04 |
ENSRNOT00000059593
|
LOC100362783
|
Uncharacterized protein C7orf61 homolog |
| chr20_+_46250363 | 0.04 |
ENSRNOT00000076522
ENSRNOT00000000334 |
Cd164
|
CD164 molecule |
| chr20_+_14577166 | 0.04 |
ENSRNOT00000085583
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr17_-_27451832 | 0.04 |
ENSRNOT00000084417
|
Riok1
|
RIO kinase 1 |
| chr4_-_21920651 | 0.03 |
ENSRNOT00000066211
|
Tmem243
|
transmembrane protein 243 |
| chr1_-_88780425 | 0.03 |
ENSRNOT00000074494
|
Sdhaf1
|
succinate dehydrogenase complex assembly factor 1 |
| chr8_-_54961265 | 0.03 |
ENSRNOT00000012434
|
Pts
|
6-pyruvoyl-tetrahydropterin synthase |
| chr1_-_67302751 | 0.03 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr4_-_174180729 | 0.03 |
ENSRNOT00000011376
|
Plcz1
|
phospholipase C, zeta 1 |
| chr20_+_41083317 | 0.03 |
ENSRNOT00000000660
|
Tspyl1
|
TSPY-like 1 |
| chr19_-_19913605 | 0.03 |
ENSRNOT00000020816
|
Cnep1r1
|
CTD nuclear envelope phosphatase 1 regulatory subunit 1 |
| chr1_-_53520788 | 0.03 |
ENSRNOT00000060121
|
Gpr31
|
G protein-coupled receptor 31 |
| chr4_-_9921610 | 0.03 |
ENSRNOT00000017018
|
Pmpcb
|
peptidase, mitochondrial processing beta subunit |
| chr10_+_55555089 | 0.03 |
ENSRNOT00000006597
|
Slc25a35
|
solute carrier family 25, member 35 |
| chr3_+_92969141 | 0.03 |
ENSRNOT00000009797
|
Apip
|
APAF1 interacting protein |
| chr6_-_1493670 | 0.03 |
ENSRNOT00000075000
|
Sult6b1
|
sulfotransferase family 6B member 1 |
| chr15_-_23969011 | 0.03 |
ENSRNOT00000014821
|
Gch1
|
GTP cyclohydrolase 1 |
| chr20_+_46667121 | 0.03 |
ENSRNOT00000089611
|
Sesn1
|
sestrin 1 |
| chr6_-_6709783 | 0.03 |
ENSRNOT00000065464
|
Cox7a2l
|
cytochrome c oxidase subunit 7A2 like |
| chr17_+_53102534 | 0.03 |
ENSRNOT00000021433
|
Mrpl32
|
mitochondrial ribosomal protein L32 |
| chr13_+_56015901 | 0.03 |
ENSRNOT00000043907
|
Dennd1b
|
DENN domain containing 1B |
| chrX_+_137197396 | 0.03 |
ENSRNOT00000082227
|
RGD1566225
|
similar to RIKEN cDNA 1700001F22 |
| chr3_-_60023913 | 0.03 |
ENSRNOT00000064662
|
Cir1
|
corepressor interacting with RBPJ, 1 |
| chr3_-_72071895 | 0.03 |
ENSRNOT00000082857
|
Selenoh
|
selenoprotein H |
| chr10_+_91126689 | 0.03 |
ENSRNOT00000004046
|
Nmt1
|
N-myristoyltransferase 1 |
| chr5_+_57267312 | 0.03 |
ENSRNOT00000011907
|
Chmp5
|
charged multivesicular body protein 5 |
| chr6_+_26517840 | 0.03 |
ENSRNOT00000031842
|
Ppm1g
|
protein phosphatase, Mg2+/Mn2+ dependent, 1G |
| chr8_+_42511520 | 0.03 |
ENSRNOT00000042974
|
Olr1259
|
olfactory receptor 1259 |
| chr1_+_147713892 | 0.03 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr5_-_164898420 | 0.03 |
ENSRNOT00000011747
|
Agtrap
|
angiotensin II receptor-associated protein |
| chr10_-_75120247 | 0.03 |
ENSRNOT00000011402
|
Lpo
|
lactoperoxidase |
| chr8_-_17524839 | 0.03 |
ENSRNOT00000081679
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chr1_-_224698514 | 0.03 |
ENSRNOT00000024234
|
Slc22a25
|
solute carrier family 22, member 25 |
| chr10_-_74001895 | 0.03 |
ENSRNOT00000005658
|
Vmp1
|
vacuole membrane protein 1 |
| chr14_-_18862407 | 0.03 |
ENSRNOT00000003823
|
Cxcl6
|
C-X-C motif chemokine ligand 6 |
| chr7_-_12399910 | 0.03 |
ENSRNOT00000021677
|
RGD1562114
|
RGD1562114 |
| chr5_-_40237591 | 0.03 |
ENSRNOT00000011393
|
Fut9
|
fucosyltransferase 9 |
| chr1_-_79690434 | 0.03 |
ENSRNOT00000057986
ENSRNOT00000057988 |
LOC102557319
|
carcinoembryonic antigen-related cell adhesion molecule 3-like |
| chrX_+_43535737 | 0.03 |
ENSRNOT00000087073
|
LOC102557137
|
E3 ubiquitin-protein ligase RNF168-like |
| chr14_+_22517774 | 0.03 |
ENSRNOT00000047655
|
Ugt2b37
|
UDP-glucuronosyltransferase 2 family, member 37 |
| chr1_-_213987053 | 0.03 |
ENSRNOT00000072774
|
LOC100911519
|
p53-induced protein with a death domain-like |
| chr10_-_110431792 | 0.03 |
ENSRNOT00000054922
|
LOC619574
|
hypothetical protein LOC619574 |
| chr8_+_39164916 | 0.03 |
ENSRNOT00000011309
|
Acrv1
|
acrosomal vesicle protein 1 |
| chr1_+_274391932 | 0.03 |
ENSRNOT00000054685
|
Rbm20
|
RNA binding motif protein 20 |
| chr2_-_53827175 | 0.03 |
ENSRNOT00000078158
|
RGD1305938
|
similar to expressed sequence AW549877 |
| chr5_-_14356692 | 0.03 |
ENSRNOT00000085654
|
Atp6v1h
|
ATPase H+ transporting V1 subunit H |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sp100
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1900673 | olefin metabolic process(GO:1900673) |
| 0.1 | 0.2 | GO:0071663 | positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) negative regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070425) negative regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070433) granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.0 | 0.1 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 0.0 | 0.1 | GO:1904612 | glucuronoside metabolic process(GO:0019389) response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.1 | GO:0042776 | mitochondrial ATP synthesis coupled proton transport(GO:0042776) |
| 0.0 | 0.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:2000422 | T-helper 1 cell lineage commitment(GO:0002296) positive regulation of mononuclear cell migration(GO:0071677) negative regulation of complement-dependent cytotoxicity(GO:1903660) regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.1 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:2000483 | negative regulation of interleukin-8 secretion(GO:2000483) |
| 0.0 | 0.1 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.0 | 0.1 | GO:1903056 | regulation of melanosome organization(GO:1903056) |
| 0.0 | 0.1 | GO:0061641 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0019401 | alditol biosynthetic process(GO:0019401) |
| 0.0 | 0.0 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.0 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0015793 | glycerol transport(GO:0015793) |
| 0.0 | 0.0 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.0 | 0.0 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:1904045 | cellular response to aldosterone(GO:1904045) |
| 0.0 | 0.0 | GO:0014916 | regulation of lung blood pressure(GO:0014916) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.0 | GO:0018201 | peptidyl-glycine modification(GO:0018201) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.0 | GO:0033185 | dolichol-phosphate-mannose synthase complex(GO:0033185) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.1 | GO:0005869 | dynactin complex(GO:0005869) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0050785 | IgE binding(GO:0019863) advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.1 | GO:0043734 | DNA-N1-methyladenine dioxygenase activity(GO:0043734) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.0 | GO:0004582 | dolichyl-phosphate beta-D-mannosyltransferase activity(GO:0004582) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:1990446 | U1 snRNP binding(GO:1990446) |
| 0.0 | 0.0 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.3 | GO:0050136 | NADH dehydrogenase (ubiquinone) activity(GO:0008137) NADH dehydrogenase (quinone) activity(GO:0050136) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0015450 | P-P-bond-hydrolysis-driven protein transmembrane transporter activity(GO:0015450) |
| 0.0 | 0.1 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |