Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
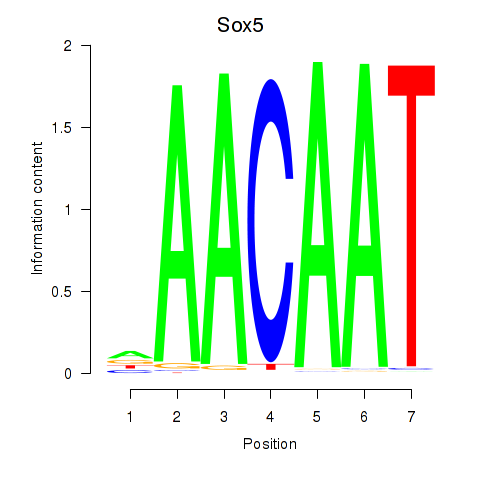
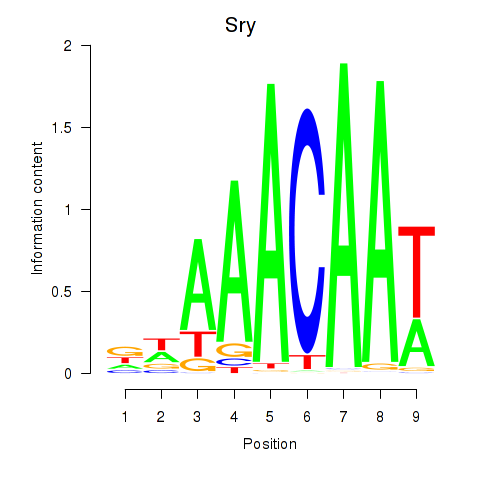
Results for Sox5_Sry
Z-value: 0.65
Transcription factors associated with Sox5_Sry
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox5
|
ENSRNOG00000027869 | SRY box 5 |
|
Sry
|
ENSRNOG00000062090 | sex determining region Y |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox5 | rn6_v1_chr4_-_178441547_178441547 | -0.88 | 4.9e-02 | Click! |
| Sry | rn6_v1_chrY_+_327166_327166 | -0.08 | 9.0e-01 | Click! |
Activity profile of Sox5_Sry motif
Sorted Z-values of Sox5_Sry motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_-_26211445 | 0.88 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr9_+_25410669 | 0.36 |
ENSRNOT00000030912
ENSRNOT00000090920 |
Tfap2b
|
transcription factor AP-2 beta |
| chr6_-_125723944 | 0.33 |
ENSRNOT00000007004
|
Fbln5
|
fibulin 5 |
| chr5_-_12563429 | 0.31 |
ENSRNOT00000059625
|
St18
|
suppression of tumorigenicity 18 |
| chr6_-_125723732 | 0.30 |
ENSRNOT00000084815
|
Fbln5
|
fibulin 5 |
| chr4_+_44774741 | 0.26 |
ENSRNOT00000086902
|
Met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr4_-_99305812 | 0.24 |
ENSRNOT00000038912
|
Rmnd5a
|
required for meiotic nuclear division 5 homolog A |
| chr11_+_61083757 | 0.23 |
ENSRNOT00000002790
|
Boc
|
BOC cell adhesion associated, oncogene regulated |
| chr14_+_17492081 | 0.21 |
ENSRNOT00000003346
|
G3bp2
|
G3BP stress granule assembly factor 2 |
| chr10_+_23661013 | 0.20 |
ENSRNOT00000076664
|
Ebf1
|
early B-cell factor 1 |
| chr4_-_82133605 | 0.19 |
ENSRNOT00000008023
|
NEWGENE_2813
|
homeobox A2 |
| chr10_-_18131562 | 0.19 |
ENSRNOT00000006676
|
Tlx3
|
T-cell leukemia, homeobox 3 |
| chr17_-_57394985 | 0.17 |
ENSRNOT00000019968
|
Epc1
|
enhancer of polycomb homolog 1 |
| chr6_-_106971250 | 0.17 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr12_-_10335499 | 0.16 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr18_-_5314511 | 0.16 |
ENSRNOT00000022637
ENSRNOT00000079682 |
Zfp521
|
zinc finger protein 521 |
| chr13_+_95589668 | 0.16 |
ENSRNOT00000005849
|
Zfp238
|
zinc finger protein 238 |
| chr10_+_86399827 | 0.15 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr20_+_5262946 | 0.15 |
ENSRNOT00000082900
|
Brd2
|
bromodomain containing 2 |
| chr16_-_19583386 | 0.14 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chr10_+_14248399 | 0.13 |
ENSRNOT00000077689
|
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr10_-_35800120 | 0.13 |
ENSRNOT00000004361
|
Maml1
|
mastermind-like transcriptional coactivator 1 |
| chr10_-_46720907 | 0.13 |
ENSRNOT00000083093
ENSRNOT00000067866 |
Tom1l2
|
target of myb1 like 2 membrane trafficking protein |
| chr15_-_24374753 | 0.13 |
ENSRNOT00000016274
|
Atg14
|
autophagy related 14 |
| chr7_-_81592206 | 0.12 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr17_+_18151982 | 0.10 |
ENSRNOT00000066447
|
Kif13a
|
kinesin family member 13A |
| chr1_+_241594565 | 0.10 |
ENSRNOT00000020123
|
Apba1
|
amyloid beta precursor protein binding family A member 1 |
| chr20_+_46429222 | 0.10 |
ENSRNOT00000076818
|
Foxo3
|
forkhead box O3 |
| chr13_+_90533365 | 0.10 |
ENSRNOT00000082469
|
Dcaf8
|
DDB1 and CUL4 associated factor 8 |
| chr4_-_123713319 | 0.10 |
ENSRNOT00000012875
ENSRNOT00000086982 |
Slc6a6
|
solute carrier family 6 member 6 |
| chr8_+_109455786 | 0.10 |
ENSRNOT00000039593
|
Msl2
|
male-specific lethal 2 homolog (Drosophila) |
| chr10_+_72909550 | 0.09 |
ENSRNOT00000004540
|
Ppm1d
|
protein phosphatase, Mg2+/Mn2+ dependent, 1D |
| chr18_+_27558089 | 0.09 |
ENSRNOT00000027499
|
Fam53c
|
family with sequence similarity 53, member C |
| chr2_-_187401786 | 0.09 |
ENSRNOT00000025624
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr6_+_110624856 | 0.09 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr10_-_16046033 | 0.09 |
ENSRNOT00000089460
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr10_-_16045835 | 0.09 |
ENSRNOT00000064832
|
Cpeb4
|
cytoplasmic polyadenylation element binding protein 4 |
| chr18_-_38088457 | 0.09 |
ENSRNOT00000077814
|
Jakmip2
|
janus kinase and microtubule interacting protein 2 |
| chr2_+_102685513 | 0.08 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chrX_-_73778595 | 0.08 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr16_-_71319449 | 0.08 |
ENSRNOT00000029284
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr16_+_20740826 | 0.08 |
ENSRNOT00000038057
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chr15_-_25505691 | 0.08 |
ENSRNOT00000074488
|
LOC100911492
|
homeobox protein OTX2-like |
| chr1_+_154606490 | 0.08 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr18_-_58097316 | 0.08 |
ENSRNOT00000088527
|
AABR07032265.1
|
|
| chr14_+_11198896 | 0.08 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr10_+_35343189 | 0.08 |
ENSRNOT00000083688
|
Mapk9
|
mitogen-activated protein kinase 9 |
| chr10_-_74298599 | 0.07 |
ENSRNOT00000007379
|
Ypel2
|
yippee-like 2 |
| chr2_-_197935567 | 0.07 |
ENSRNOT00000085404
|
Rprd2
|
regulation of nuclear pre-mRNA domain containing 2 |
| chr4_+_161696903 | 0.07 |
ENSRNOT00000055889
|
LOC100362909
|
rCG30099-like |
| chr7_-_59547174 | 0.07 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr3_-_94833406 | 0.07 |
ENSRNOT00000049695
|
RGD1560017
|
similar to Ac2-210 |
| chr1_-_280338813 | 0.07 |
ENSRNOT00000011743
|
Vax1
|
ventral anterior homeobox 1 |
| chr3_+_80676820 | 0.07 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr13_-_83425641 | 0.07 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr13_+_106463368 | 0.07 |
ENSRNOT00000003489
|
Esrrg
|
estrogen-related receptor gamma |
| chr19_-_26053762 | 0.06 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr9_+_16647922 | 0.06 |
ENSRNOT00000031625
|
Klc4
|
kinesin light chain 4 |
| chrX_+_63343724 | 0.06 |
ENSRNOT00000076530
ENSRNOT00000008558 |
Klhl15
|
kelch-like family member 15 |
| chr1_-_261090437 | 0.06 |
ENSRNOT00000072055
|
Frat2
|
FRAT2, WNT signaling pathway regulator |
| chrX_+_6273733 | 0.06 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr2_-_96509424 | 0.06 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr4_+_152892388 | 0.06 |
ENSRNOT00000056198
ENSRNOT00000090218 ENSRNOT00000075895 |
Kdm5a
|
lysine demethylase 5A |
| chr2_-_231521052 | 0.06 |
ENSRNOT00000089534
ENSRNOT00000080470 ENSRNOT00000084756 |
Ank2
|
ankyrin 2 |
| chr10_+_71536533 | 0.06 |
ENSRNOT00000088138
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr6_-_67084234 | 0.06 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr11_+_9642365 | 0.05 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr16_+_23668595 | 0.05 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr2_-_63166509 | 0.05 |
ENSRNOT00000018246
|
Cdh6
|
cadherin 6 |
| chrX_-_73766885 | 0.05 |
ENSRNOT00000091325
|
Rlim
|
ring finger protein, LIM domain interacting |
| chr9_+_16647598 | 0.05 |
ENSRNOT00000087413
|
Klc4
|
kinesin light chain 4 |
| chr16_+_46731403 | 0.05 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr7_-_129724303 | 0.05 |
ENSRNOT00000006034
|
Brd1
|
bromodomain containing 1 |
| chr3_+_116899878 | 0.05 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr6_+_147876237 | 0.05 |
ENSRNOT00000056649
|
Tmem196
|
transmembrane protein 196 |
| chr9_-_43139870 | 0.05 |
ENSRNOT00000021956
|
Sema4c
|
semaphorin 4C |
| chr7_-_138483612 | 0.05 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr13_+_98615287 | 0.05 |
ENSRNOT00000004032
|
Itpkb
|
inositol-trisphosphate 3-kinase B |
| chr13_+_76943633 | 0.05 |
ENSRNOT00000076908
|
Rfwd2
|
ring finger and WD repeat domain 2 |
| chr11_-_84633504 | 0.05 |
ENSRNOT00000052120
|
Klhl24
|
kelch-like family member 24 |
| chr10_+_11206226 | 0.05 |
ENSRNOT00000006979
|
Tfap4
|
transcription factor AP-4 |
| chr14_+_2613406 | 0.05 |
ENSRNOT00000000083
|
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr4_-_115516296 | 0.05 |
ENSRNOT00000019399
|
Paip2b
|
poly(A) binding protein interacting protein 2B |
| chr14_+_81819415 | 0.05 |
ENSRNOT00000018515
ENSRNOT00000075863 ENSRNOT00000077060 |
Mxd4
|
Max dimerization protein 4 |
| chr14_+_11199404 | 0.05 |
ENSRNOT00000003106
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr9_+_67234303 | 0.05 |
ENSRNOT00000050179
|
Abi2
|
abl-interactor 2 |
| chr17_-_18421861 | 0.05 |
ENSRNOT00000060500
ENSRNOT00000036560 |
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr2_+_198006316 | 0.05 |
ENSRNOT00000081383
ENSRNOT00000056289 |
Aph1a
|
aph-1 homolog A, gamma secretase subunit |
| chr6_-_136436620 | 0.05 |
ENSRNOT00000067118
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr1_-_126211439 | 0.05 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr10_+_14248710 | 0.04 |
ENSRNOT00000020733
|
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr4_-_119131202 | 0.04 |
ENSRNOT00000011675
|
Antxr1
|
anthrax toxin receptor 1 |
| chr6_-_136436818 | 0.04 |
ENSRNOT00000082600
|
Ppp1r13b
|
protein phosphatase 1, regulatory subunit 13B |
| chr4_+_155321553 | 0.04 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr4_+_62221011 | 0.04 |
ENSRNOT00000041264
|
Cald1
|
caldesmon 1 |
| chr3_+_14990652 | 0.04 |
ENSRNOT00000090735
|
Dab2ip
|
DAB2 interacting protein |
| chr1_-_93949187 | 0.04 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr7_+_142776252 | 0.04 |
ENSRNOT00000008673
|
Acvrl1
|
activin A receptor like type 1 |
| chr20_+_34894419 | 0.04 |
ENSRNOT00000000471
|
Asf1a
|
anti-silencing function 1A histone chaperone |
| chr5_-_50684409 | 0.04 |
ENSRNOT00000013181
|
RGD1359108
|
similar to RIKEN cDNA 3110043O21 |
| chr7_+_59326518 | 0.04 |
ENSRNOT00000085231
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr2_-_172361779 | 0.04 |
ENSRNOT00000085876
|
Schip1
|
schwannomin interacting protein 1 |
| chr3_-_94418711 | 0.04 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chrX_+_62727755 | 0.04 |
ENSRNOT00000077055
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr10_+_66732390 | 0.04 |
ENSRNOT00000089538
|
Nf1
|
neurofibromin 1 |
| chr1_-_40921508 | 0.04 |
ENSRNOT00000026433
|
Zbtb2
|
zinc finger and BTB domain containing 2 |
| chr16_-_71319052 | 0.04 |
ENSRNOT00000050980
|
Fgfr1
|
Fibroblast growth factor receptor 1 |
| chr8_+_119030875 | 0.04 |
ENSRNOT00000028458
|
Myl3
|
myosin light chain 3 |
| chr10_+_4945911 | 0.03 |
ENSRNOT00000003420
|
Prm1
|
protamine 1 |
| chr4_+_62220736 | 0.03 |
ENSRNOT00000086377
|
Cald1
|
caldesmon 1 |
| chr2_-_177924970 | 0.03 |
ENSRNOT00000029340
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr8_+_128824508 | 0.03 |
ENSRNOT00000025343
|
Mobp
|
myelin-associated oligodendrocyte basic protein |
| chr4_+_158088505 | 0.03 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr1_-_156327352 | 0.03 |
ENSRNOT00000074282
|
LOC100911776
|
coiled-coil domain-containing protein 89-like |
| chr9_+_62291809 | 0.03 |
ENSRNOT00000090621
|
Plcl1
|
phospholipase C-like 1 |
| chr5_+_113725717 | 0.03 |
ENSRNOT00000032248
|
Tek
|
TEK receptor tyrosine kinase |
| chr9_+_46657575 | 0.03 |
ENSRNOT00000083322
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr9_+_2190915 | 0.03 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr1_-_13395370 | 0.03 |
ENSRNOT00000085519
|
Ccdc28a
|
coiled-coil domain containing 28A |
| chr6_+_29061618 | 0.03 |
ENSRNOT00000092223
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr15_+_83703791 | 0.03 |
ENSRNOT00000090637
|
Klf5
|
Kruppel-like factor 5 |
| chr14_+_10786399 | 0.03 |
ENSRNOT00000003006
|
Lin54
|
lin-54 DREAM MuvB core complex component |
| chr3_+_148510779 | 0.03 |
ENSRNOT00000012156
|
Xkr7
|
XK related 7 |
| chr7_+_59349745 | 0.03 |
ENSRNOT00000085334
|
Ptprb
|
protein tyrosine phosphatase, receptor type, B |
| chr8_+_94686938 | 0.03 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr4_+_157524423 | 0.03 |
ENSRNOT00000036654
|
Zfp384
|
zinc finger protein 384 |
| chr3_-_91370167 | 0.03 |
ENSRNOT00000006291
|
Prr5l
|
proline rich 5 like |
| chr3_-_71845232 | 0.03 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr4_-_168517177 | 0.03 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr13_+_60435946 | 0.03 |
ENSRNOT00000004342
|
B3galt2
|
Beta-1,3-galactosyltransferase 2 |
| chr7_+_142776580 | 0.03 |
ENSRNOT00000081047
|
Acvrl1
|
activin A receptor like type 1 |
| chr8_+_48925604 | 0.03 |
ENSRNOT00000077445
|
Ddx6
|
DEAD-box helicase 6 |
| chr10_+_43768708 | 0.03 |
ENSRNOT00000086218
|
Sh3bp5l
|
SH3 binding domain protein 5 like |
| chr6_-_105518725 | 0.02 |
ENSRNOT00000009693
ENSRNOT00000083488 |
Map3k9
|
mitogen-activated protein kinase kinase kinase 9 |
| chr2_+_189615948 | 0.02 |
ENSRNOT00000092144
|
Crtc2
|
CREB regulated transcription coactivator 2 |
| chr8_-_71337746 | 0.02 |
ENSRNOT00000021554
|
Zfp609
|
zinc finger protein 609 |
| chr4_+_118207862 | 0.02 |
ENSRNOT00000085787
ENSRNOT00000023103 |
Tia1
|
TIA1 cytotoxic granule-associated RNA binding protein |
| chr9_-_16647458 | 0.02 |
ENSRNOT00000024380
|
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chr10_+_39850818 | 0.02 |
ENSRNOT00000012671
|
Fnip1
|
folliculin interacting protein 1 |
| chr8_+_117574161 | 0.02 |
ENSRNOT00000077078
|
Ip6k2
|
inositol hexakisphosphate kinase 2 |
| chr14_+_8080565 | 0.02 |
ENSRNOT00000092395
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr13_+_89805962 | 0.02 |
ENSRNOT00000074035
|
Tstd1
|
thiosulfate sulfurtransferase like domain containing 1 |
| chr10_+_67862054 | 0.02 |
ENSRNOT00000031746
|
Cdk5r1
|
cyclin-dependent kinase 5 regulatory subunit 1 |
| chr2_+_257633425 | 0.02 |
ENSRNOT00000071770
|
Zzz3
|
zinc finger, ZZ-type containing 3 |
| chr6_-_107080524 | 0.02 |
ENSRNOT00000011662
|
Zfyve1
|
zinc finger FYVE-type containing 1 |
| chr15_+_39779648 | 0.02 |
ENSRNOT00000084505
|
Cab39l
|
calcium binding protein 39-like |
| chr9_-_55256340 | 0.02 |
ENSRNOT00000028907
|
Sdpr
|
serum deprivation response |
| chr3_-_110324084 | 0.02 |
ENSRNOT00000010381
|
Bmf
|
Bcl2 modifying factor |
| chrX_+_9436707 | 0.02 |
ENSRNOT00000004187
|
Cask
|
calcium/calmodulin dependent serine protein kinase |
| chr16_-_21274688 | 0.02 |
ENSRNOT00000027953
|
Tssk6
|
testis-specific serine kinase 6 |
| chr3_-_168018410 | 0.02 |
ENSRNOT00000087579
|
Bcas1
|
breast carcinoma amplified sequence 1 |
| chr10_+_95642640 | 0.01 |
ENSRNOT00000029782
|
RGD1359290
|
Ribosomal_L22 domain containing protein RGD1359290 |
| chr18_+_79406381 | 0.01 |
ENSRNOT00000022303
ENSRNOT00000058295 ENSRNOT00000058296 ENSRNOT00000022280 |
Mbp
|
myelin basic protein |
| chr6_+_27768943 | 0.01 |
ENSRNOT00000015820
|
Kif3c
|
kinesin family member 3C |
| chr9_-_5329305 | 0.01 |
ENSRNOT00000078055
|
Slc5a7
|
solute carrier family 5 member 7 |
| chr18_+_3995921 | 0.01 |
ENSRNOT00000091424
|
Ttc39c
|
tetratricopeptide repeat domain 39C |
| chr4_-_125929002 | 0.01 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr10_-_14247886 | 0.01 |
ENSRNOT00000020504
|
Nubp2
|
nucleotide binding protein 2 |
| chr1_-_137359072 | 0.01 |
ENSRNOT00000014553
|
Klhl25
|
kelch-like family member 25 |
| chr1_+_12823363 | 0.01 |
ENSRNOT00000086790
|
Cited2
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 |
| chr1_-_8885967 | 0.01 |
ENSRNOT00000016102
|
Gje1
|
gap junction protein, epsilon 1 |
| chr4_+_7122890 | 0.01 |
ENSRNOT00000076339
ENSRNOT00000076418 ENSRNOT00000014718 |
Abcf2
|
ATP binding cassette subfamily F member 2 |
| chr1_+_174702373 | 0.01 |
ENSRNOT00000013733
|
Zfp143
|
zinc finger protein 143 |
| chr18_+_3887419 | 0.01 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr5_-_48504511 | 0.01 |
ENSRNOT00000010271
|
Pnrc1
|
proline-rich nuclear receptor coactivator 1 |
| chrX_+_128493614 | 0.01 |
ENSRNOT00000044240
|
Stag2
|
stromal antigen 2 |
| chr3_-_147865393 | 0.01 |
ENSRNOT00000009852
|
Sox12
|
SRY box 12 |
| chr4_-_170117325 | 0.01 |
ENSRNOT00000074198
|
LOC100912564
|
histone H4-like |
| chr4_-_87111025 | 0.01 |
ENSRNOT00000018442
|
Kbtbd2
|
kelch repeat and BTB domain containing 2 |
| chr1_+_259926537 | 0.01 |
ENSRNOT00000073537
|
NEWGENE_1306399
|
cyclin J |
| chr9_-_92405117 | 0.01 |
ENSRNOT00000079452
|
Trip12
|
thyroid hormone receptor interactor 12 |
| chr4_-_129569470 | 0.00 |
ENSRNOT00000008893
|
Uba3
|
ubiquitin-like modifier activating enzyme 3 |
| chrX_+_63343546 | 0.00 |
ENSRNOT00000076315
|
Klhl15
|
kelch-like family member 15 |
| chrX_+_40363646 | 0.00 |
ENSRNOT00000010135
|
Sms
|
spermine synthase |
| chr10_+_35872619 | 0.00 |
ENSRNOT00000059190
|
Hnrnph1
|
heterogeneous nuclear ribonucleoprotein H1 |
| chrX_-_62690806 | 0.00 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr16_-_47535358 | 0.00 |
ENSRNOT00000040731
|
Cldn22
|
claudin 22 |
| chr3_+_57286892 | 0.00 |
ENSRNOT00000039065
|
LOC103690018
|
Golgi reassembly-stacking protein 2-like |
| chr2_+_77868412 | 0.00 |
ENSRNOT00000065897
ENSRNOT00000014022 |
Myo10
|
myosin X |
| chr3_-_160038078 | 0.00 |
ENSRNOT00000013445
|
Serinc3
|
serine incorporator 3 |
| chr13_-_68858781 | 0.00 |
ENSRNOT00000065944
ENSRNOT00000092053 |
Rnf2
|
ring finger protein 2 |
| chr4_-_55398941 | 0.00 |
ENSRNOT00000075324
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr1_+_260798239 | 0.00 |
ENSRNOT00000036791
|
Lcor
|
ligand dependent nuclear receptor corepressor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox5_Sry
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.1 | 0.4 | GO:0060665 | regulation of branching involved in salivary gland morphogenesis by mesenchymal-epithelial signaling(GO:0060665) |
| 0.1 | 0.4 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.1 | 0.2 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.1 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0060926 | cardiac pacemaker cell development(GO:0060926) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.4 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.2 | GO:1904646 | cellular response to beta-amyloid(GO:1904646) |
| 0.0 | 0.1 | GO:0021823 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) |
| 0.0 | 0.2 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.0 | GO:1901726 | negative regulation of histone deacetylase activity(GO:1901726) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:2001295 | multicellular organismal protein metabolic process(GO:0044268) malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0071630 | nucleus-associated proteasomal ubiquitin-dependent protein catabolic process(GO:0071630) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
| 0.0 | 0.9 | GO:0031103 | axon regeneration(GO:0031103) |
| 0.0 | 0.1 | GO:0021960 | anterior commissure morphogenesis(GO:0021960) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.2 | GO:0002087 | regulation of respiratory gaseous exchange by neurological system process(GO:0002087) |
| 0.0 | 0.1 | GO:0033030 | negative regulation of neutrophil apoptotic process(GO:0033030) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.1 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.1 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0030478 | actin cap(GO:0030478) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030977 | taurine binding(GO:0030977) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.3 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0098821 | activin receptor activity, type I(GO:0016361) BMP receptor activity(GO:0098821) |
| 0.0 | 0.1 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0043274 | phospholipase binding(GO:0043274) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.1 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.0 | 0.1 | REACTOME SIGNALING BY ACTIVATED POINT MUTANTS OF FGFR1 | Genes involved in Signaling by activated point mutants of FGFR1 |