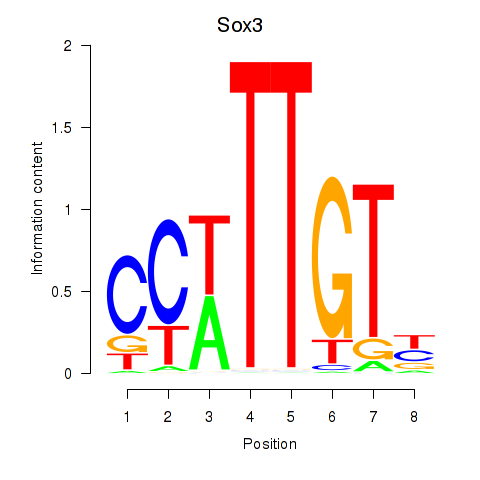
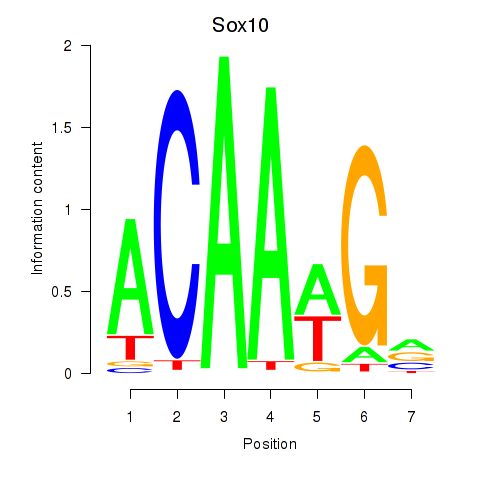
Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Sox3_Sox10
Z-value: 0.32
Transcription factors associated with Sox3_Sox10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox10
|
ENSRNOG00000011305 | SRY box 10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox10 | rn6_v1_chr7_-_120403523_120403523 | -0.44 | 4.6e-01 | Click! |
Activity profile of Sox3_Sox10 motif
Sorted Z-values of Sox3_Sox10 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_155160481 | 0.14 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr12_-_13668515 | 0.14 |
ENSRNOT00000086847
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr10_-_88670430 | 0.14 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr18_+_30562178 | 0.11 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr3_-_152382529 | 0.10 |
ENSRNOT00000027076
|
Scand1
|
SCAN domain-containing 1 |
| chr20_+_3558827 | 0.10 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr9_+_46657575 | 0.09 |
ENSRNOT00000083322
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chrX_+_80213332 | 0.09 |
ENSRNOT00000042827
|
Sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr10_-_85435016 | 0.09 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr15_-_43542939 | 0.09 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr14_-_81399353 | 0.09 |
ENSRNOT00000018340
|
Add1
|
adducin 1 |
| chr9_+_49479023 | 0.08 |
ENSRNOT00000050922
ENSRNOT00000077111 |
Pou3f3
|
POU class 3 homeobox 3 |
| chrX_+_62754634 | 0.08 |
ENSRNOT00000016669
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr18_+_30895831 | 0.08 |
ENSRNOT00000026985
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr7_-_12275609 | 0.08 |
ENSRNOT00000086061
|
Apc2
|
APC2, WNT signaling pathway regulator |
| chr2_-_226779266 | 0.07 |
ENSRNOT00000019244
|
Fnbp1l
|
formin binding protein 1-like |
| chr14_+_81819799 | 0.07 |
ENSRNOT00000076840
|
Mxd4
|
Max dimerization protein 4 |
| chr2_-_117666683 | 0.07 |
ENSRNOT00000015479
|
AABR07009931.1
|
|
| chr8_+_128824508 | 0.07 |
ENSRNOT00000025343
|
Mobp
|
myelin-associated oligodendrocyte basic protein |
| chr16_+_83522162 | 0.07 |
ENSRNOT00000057386
|
Col4a1
|
collagen type IV alpha 1 chain |
| chr4_-_168517177 | 0.07 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chrX_-_115175299 | 0.07 |
ENSRNOT00000074322
|
Dcx
|
doublecortin |
| chr1_-_199177365 | 0.07 |
ENSRNOT00000041840
|
Ctf2
|
cardiotrophin 2 |
| chr17_+_70684134 | 0.07 |
ENSRNOT00000025731
ENSRNOT00000068354 |
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr2_+_190003223 | 0.06 |
ENSRNOT00000015712
|
S100a5
|
S100 calcium binding protein A5 |
| chr20_+_4993560 | 0.06 |
ENSRNOT00000081628
ENSRNOT00000087861 ENSRNOT00000001160 |
Vars
|
valyl-tRNA synthetase |
| chr8_-_62987182 | 0.06 |
ENSRNOT00000070885
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr5_-_153625869 | 0.06 |
ENSRNOT00000024464
|
Clic4
|
chloride intracellular channel 4 |
| chr10_+_109773890 | 0.06 |
ENSRNOT00000071823
|
AC131537.1
|
|
| chr1_-_228753422 | 0.06 |
ENSRNOT00000028626
|
Dtx4
|
deltex E3 ubiquitin ligase 4 |
| chr17_-_57394985 | 0.06 |
ENSRNOT00000019968
|
Epc1
|
enhancer of polycomb homolog 1 |
| chr3_+_152571121 | 0.06 |
ENSRNOT00000087289
ENSRNOT00000080543 ENSRNOT00000083476 |
Epb41l1
|
erythrocyte membrane protein band 4.1-like 1 |
| chr10_-_47453442 | 0.06 |
ENSRNOT00000050061
|
Usp22
|
ubiquitin specific peptidase 22 |
| chr3_+_162357356 | 0.06 |
ENSRNOT00000030119
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr1_-_20155960 | 0.06 |
ENSRNOT00000061389
|
Samd3
|
sterile alpha motif domain containing 3 |
| chr11_+_61531416 | 0.06 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr17_+_70684539 | 0.06 |
ENSRNOT00000025700
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr10_+_62674561 | 0.06 |
ENSRNOT00000019946
ENSRNOT00000056110 |
Ankrd13b
|
ankyrin repeat domain 13B |
| chr2_+_22909569 | 0.05 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr9_-_100253609 | 0.05 |
ENSRNOT00000036061
|
Kif1a
|
kinesin family member 1A |
| chr9_-_60923706 | 0.05 |
ENSRNOT00000047517
|
AABR07067749.1
|
|
| chr4_-_125929002 | 0.05 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr8_+_70884491 | 0.05 |
ENSRNOT00000072939
|
Ubap1l
|
ubiquitin associated protein 1-like |
| chr5_-_147412705 | 0.05 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr19_+_755460 | 0.05 |
ENSRNOT00000076560
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr12_-_40822159 | 0.04 |
ENSRNOT00000001826
|
Hectd4
|
HECT domain E3 ubiquitin protein ligase 4 |
| chr20_+_40769586 | 0.04 |
ENSRNOT00000001079
|
Fabp7
|
fatty acid binding protein 7 |
| chr13_-_51076852 | 0.04 |
ENSRNOT00000078993
|
Adora1
|
adenosine A1 receptor |
| chr3_+_151126591 | 0.04 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr12_+_27155587 | 0.04 |
ENSRNOT00000044800
|
AABR07035916.1
|
|
| chr8_+_59457018 | 0.04 |
ENSRNOT00000017900
|
Ireb2
|
iron responsive element binding protein 2 |
| chr9_-_28972835 | 0.04 |
ENSRNOT00000086967
ENSRNOT00000079684 |
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr2_-_31753528 | 0.04 |
ENSRNOT00000075057
|
Pik3r1
|
phosphoinositide-3-kinase regulatory subunit 1 |
| chr10_-_90393317 | 0.04 |
ENSRNOT00000028563
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr18_+_30381322 | 0.04 |
ENSRNOT00000027216
|
Pcdhb3
|
protocadherin beta 3 |
| chr1_+_259926537 | 0.04 |
ENSRNOT00000073537
|
NEWGENE_1306399
|
cyclin J |
| chr1_+_140998240 | 0.04 |
ENSRNOT00000023506
ENSRNOT00000090897 |
Abhd2
|
abhydrolase domain containing 2 |
| chr17_-_77687456 | 0.04 |
ENSRNOT00000045765
ENSRNOT00000081645 |
Frmd4a
|
FERM domain containing 4A |
| chr2_+_95008477 | 0.04 |
ENSRNOT00000015327
|
Tpd52
|
tumor protein D52 |
| chr11_-_1836897 | 0.04 |
ENSRNOT00000050533
|
Zfp654
|
zinc finger protein 654 |
| chr9_+_73433252 | 0.04 |
ENSRNOT00000092540
|
Map2
|
microtubule-associated protein 2 |
| chr2_-_226779440 | 0.04 |
ENSRNOT00000055669
|
Fnbp1l
|
formin binding protein 1-like |
| chr16_-_19894591 | 0.04 |
ENSRNOT00000085940
|
Ano8
|
anoctamin 8 |
| chr20_-_2701637 | 0.04 |
ENSRNOT00000049667
|
Hspa1a
|
heat shock protein family A (Hsp70) member 1A |
| chr12_+_38303624 | 0.04 |
ENSRNOT00000001668
|
Zcchc8
|
zinc finger CCHC-type containing 8 |
| chr18_+_15467870 | 0.04 |
ENSRNOT00000091696
|
B4galt6
|
beta-1,4-galactosyltransferase 6 |
| chr1_+_81372650 | 0.03 |
ENSRNOT00000088829
|
Zfp428
|
zinc finger protein 428 |
| chr15_-_52399074 | 0.03 |
ENSRNOT00000018440
|
Xpo7
|
exportin 7 |
| chr3_+_164665532 | 0.03 |
ENSRNOT00000014309
|
Ptpn1
|
protein tyrosine phosphatase, non-receptor type 1 |
| chr12_+_40018937 | 0.03 |
ENSRNOT00000001697
|
Cux2
|
cut-like homeobox 2 |
| chr10_-_17075139 | 0.03 |
ENSRNOT00000039398
|
Neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chrX_-_73778595 | 0.03 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr17_-_4454701 | 0.03 |
ENSRNOT00000080750
ENSRNOT00000066950 |
Dapk1
|
death associated protein kinase 1 |
| chr14_+_11198896 | 0.03 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr15_+_10120206 | 0.03 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chrX_+_88298266 | 0.03 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr6_+_73553210 | 0.03 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr17_+_63635086 | 0.03 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr1_-_18058055 | 0.03 |
ENSRNOT00000020988
|
Ptprk
|
protein tyrosine phosphatase, receptor type, K |
| chr2_+_154604832 | 0.03 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr10_+_78168715 | 0.03 |
ENSRNOT00000072294
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr20_+_3552929 | 0.03 |
ENSRNOT00000083223
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr9_+_14551758 | 0.03 |
ENSRNOT00000017157
|
Nfya
|
nuclear transcription factor Y subunit alpha |
| chr4_+_158088505 | 0.03 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr8_-_125544427 | 0.03 |
ENSRNOT00000041548
ENSRNOT00000039904 ENSRNOT00000036642 |
Rbms3
|
RNA binding motif, single stranded interacting protein 3 |
| chr18_-_69671199 | 0.03 |
ENSRNOT00000082484
|
Smad4
|
SMAD family member 4 |
| chr18_+_30890869 | 0.03 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr18_+_14757679 | 0.03 |
ENSRNOT00000071364
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr3_-_9448167 | 0.03 |
ENSRNOT00000012617
|
Abl1
|
ABL proto-oncogene 1, non-receptor tyrosine kinase |
| chrX_+_62727755 | 0.03 |
ENSRNOT00000077055
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr7_+_64672722 | 0.03 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr15_-_62200837 | 0.03 |
ENSRNOT00000017599
|
Pcdh8
|
protocadherin 8 |
| chr3_-_148312791 | 0.03 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr5_+_49311030 | 0.03 |
ENSRNOT00000010850
|
Cnr1
|
cannabinoid receptor 1 |
| chr8_+_129617812 | 0.03 |
ENSRNOT00000079085
|
Ctnnb1
|
catenin beta 1 |
| chr18_+_30435119 | 0.03 |
ENSRNOT00000027190
|
Pcdhb8
|
protocadherin beta 8 |
| chr17_+_13670520 | 0.03 |
ENSRNOT00000019442
|
Shc3
|
SHC adaptor protein 3 |
| chr12_-_35979193 | 0.03 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr16_+_31734944 | 0.03 |
ENSRNOT00000059673
|
Palld
|
palladin, cytoskeletal associated protein |
| chr7_+_99954492 | 0.03 |
ENSRNOT00000005885
|
Trib1
|
tribbles pseudokinase 1 |
| chr8_-_61919874 | 0.03 |
ENSRNOT00000064830
|
RGD1305464
|
similar to human chromosome 15 open reading frame 39 |
| chr5_-_86696388 | 0.03 |
ENSRNOT00000007812
|
Megf9
|
multiple EGF-like-domains 9 |
| chr17_-_44527801 | 0.03 |
ENSRNOT00000089643
|
Hist1h2bk
|
histone cluster 1 H2B family member k |
| chr10_+_71536533 | 0.03 |
ENSRNOT00000088138
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr20_-_6257604 | 0.03 |
ENSRNOT00000092489
|
Stk38
|
serine/threonine kinase 38 |
| chr20_+_47494424 | 0.03 |
ENSRNOT00000087718
|
Sec63
|
SEC63 homolog, protein translocation regulator |
| chr15_+_44441856 | 0.03 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr9_-_85528860 | 0.03 |
ENSRNOT00000063834
ENSRNOT00000088963 |
Wdfy1
|
WD repeat and FYVE domain containing 1 |
| chr8_+_116325286 | 0.03 |
ENSRNOT00000017461
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr6_-_147172022 | 0.03 |
ENSRNOT00000080675
|
Itgb8
|
integrin subunit beta 8 |
| chr6_+_147876237 | 0.03 |
ENSRNOT00000056649
|
Tmem196
|
transmembrane protein 196 |
| chr16_+_11032531 | 0.03 |
ENSRNOT00000078390
|
Wapl
|
WAPL cohesin release factor |
| chrX_-_70460536 | 0.03 |
ENSRNOT00000076824
|
Pdzd11
|
PDZ domain containing 11 |
| chr9_+_6966908 | 0.03 |
ENSRNOT00000072796
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr2_-_187820952 | 0.03 |
ENSRNOT00000092762
|
Sema4a
|
semaphorin 4A |
| chr6_+_48866601 | 0.03 |
ENSRNOT00000077321
ENSRNOT00000079891 |
Pxdn
|
peroxidasin |
| chr2_+_8732932 | 0.03 |
ENSRNOT00000072849
|
Arrdc3
|
arrestin domain containing 3 |
| chr3_+_176102351 | 0.03 |
ENSRNOT00000013105
|
Col9a3
|
collagen type IX alpha 3 chain |
| chr5_-_139748489 | 0.03 |
ENSRNOT00000078741
|
Nfyc
|
nuclear transcription factor Y subunit gamma |
| chr1_-_82279145 | 0.02 |
ENSRNOT00000057433
|
Cxcl17
|
C-X-C motif chemokine ligand 17 |
| chr12_-_25638797 | 0.02 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr9_-_28973246 | 0.02 |
ENSRNOT00000091865
ENSRNOT00000015453 |
Rims1
|
regulating synaptic membrane exocytosis 1 |
| chr14_+_2613406 | 0.02 |
ENSRNOT00000000083
|
Tmed5
|
transmembrane p24 trafficking protein 5 |
| chr8_-_6203515 | 0.02 |
ENSRNOT00000087278
ENSRNOT00000031189 ENSRNOT00000008074 ENSRNOT00000085285 ENSRNOT00000007866 |
Yap1
|
yes-associated protein 1 |
| chr7_-_136853154 | 0.02 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr4_-_183544850 | 0.02 |
ENSRNOT00000071407
|
Dennd5b
|
DENN/MADD domain containing 5B |
| chrX_-_29648359 | 0.02 |
ENSRNOT00000086721
ENSRNOT00000006777 |
Gpm6b
|
glycoprotein m6b |
| chr13_+_52976507 | 0.02 |
ENSRNOT00000090599
ENSRNOT00000011324 |
Kif21b
|
kinesin family member 21B |
| chr13_-_107831014 | 0.02 |
ENSRNOT00000003684
|
Kcnk2
|
potassium two pore domain channel subfamily K member 2 |
| chr8_+_116054465 | 0.02 |
ENSRNOT00000040056
|
Cish
|
cytokine inducible SH2-containing protein |
| chr15_+_86243148 | 0.02 |
ENSRNOT00000084471
ENSRNOT00000090727 |
Lmo7
|
LIM domain 7 |
| chr1_+_277459200 | 0.02 |
ENSRNOT00000086008
|
AABR07007023.1
|
|
| chr17_-_13393243 | 0.02 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr1_-_213981317 | 0.02 |
ENSRNOT00000086238
ENSRNOT00000054869 |
Slc25a22
|
solute carrier family 25 member 22 |
| chr10_-_51576376 | 0.02 |
ENSRNOT00000004829
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr17_+_70684340 | 0.02 |
ENSRNOT00000051067
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr16_+_74752655 | 0.02 |
ENSRNOT00000029266
|
Ckap2
|
cytoskeleton associated protein 2 |
| chr8_-_116893057 | 0.02 |
ENSRNOT00000082113
|
Bsn
|
bassoon (presynaptic cytomatrix protein) |
| chrX_-_15553720 | 0.02 |
ENSRNOT00000012646
|
Gripap1
|
GRIP1 associated protein 1 |
| chr1_-_68269117 | 0.02 |
ENSRNOT00000072367
|
LOC100911224
|
zinc finger protein interacting with ribonucleoprotein K-like |
| chr19_+_740028 | 0.02 |
ENSRNOT00000076604
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr1_-_103256823 | 0.02 |
ENSRNOT00000018860
|
Ptpn5
|
protein tyrosine phosphatase, non-receptor type 5 |
| chr18_+_30880020 | 0.02 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr13_-_111581018 | 0.02 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr10_-_14788617 | 0.02 |
ENSRNOT00000043626
|
Cacna1h
|
calcium voltage-gated channel subunit alpha1 H |
| chr19_-_55490426 | 0.02 |
ENSRNOT00000081800
|
Cbfa2t3
|
CBFA2/RUNX1 translocation partner 3 |
| chr18_+_79773608 | 0.02 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr4_-_150616895 | 0.02 |
ENSRNOT00000073562
|
Ankrd26
|
ankyrin repeat domain 26 |
| chr18_+_30909490 | 0.02 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr2_-_23909016 | 0.02 |
ENSRNOT00000084044
|
Scamp1
|
secretory carrier membrane protein 1 |
| chr5_-_58484900 | 0.02 |
ENSRNOT00000012386
|
Fam214b
|
family with sequence similarity 214, member B |
| chr7_+_120891940 | 0.02 |
ENSRNOT00000018787
|
Cby1
|
chibby family member 1, beta catenin antagonist |
| chr15_+_105851542 | 0.02 |
ENSRNOT00000086959
|
Rap2a
|
RAS related protein 2a |
| chr20_+_3582743 | 0.02 |
ENSRNOT00000001103
|
Gtf2h4
|
general transcription factor 2H subunit 4 |
| chr14_-_37770059 | 0.02 |
ENSRNOT00000029721
|
Slc10a4
|
solute carrier family 10, member 4 |
| chr11_+_86512797 | 0.02 |
ENSRNOT00000051680
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr1_-_126211439 | 0.02 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr16_+_54319377 | 0.02 |
ENSRNOT00000090266
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr7_+_12652415 | 0.02 |
ENSRNOT00000031315
|
Plppr3
|
phospholipid phosphatase related 3 |
| chr10_+_10644572 | 0.02 |
ENSRNOT00000004026
|
Ppl
|
periplakin |
| chr8_-_111965889 | 0.02 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr9_-_61810417 | 0.02 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr6_+_80159364 | 0.02 |
ENSRNOT00000005439
|
Pnn
|
pinin, desmosome associated protein |
| chr14_+_63095720 | 0.02 |
ENSRNOT00000006071
|
Ppargc1a
|
PPARG coactivator 1 alpha |
| chr7_-_11824742 | 0.02 |
ENSRNOT00000051467
|
Dot1l
|
DOT1 like histone lysine methyltransferase |
| chr18_+_4084228 | 0.02 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chr3_-_80601410 | 0.02 |
ENSRNOT00000022841
|
Atg13
|
autophagy related 13 |
| chr1_-_176607466 | 0.02 |
ENSRNOT00000022984
|
Galnt18
|
polypeptide N-acetylgalactosaminyltransferase 18 |
| chr12_+_28381982 | 0.02 |
ENSRNOT00000076101
|
Wbscr17
|
Williams-Beuren syndrome chromosome region 17 |
| chr3_-_160853650 | 0.02 |
ENSRNOT00000018844
|
Matn4
|
matrilin 4 |
| chr2_+_122368265 | 0.02 |
ENSRNOT00000078321
|
Atp11b
|
ATPase phospholipid transporting 11B (putative) |
| chr19_+_9024451 | 0.02 |
ENSRNOT00000073744
|
LOC100910721
|
60S ribosomal protein L26-like |
| chr13_+_79269973 | 0.02 |
ENSRNOT00000003969
|
Tnfsf4
|
tumor necrosis factor superfamily member 4 |
| chr7_+_34952011 | 0.02 |
ENSRNOT00000077666
|
Fgd6
|
FYVE, RhoGEF and PH domain containing 6 |
| chrX_-_26376467 | 0.02 |
ENSRNOT00000051655
ENSRNOT00000036502 |
Arhgap6
|
Rho GTPase activating protein 6 |
| chr14_+_64686793 | 0.02 |
ENSRNOT00000005894
ENSRNOT00000036646 |
Adgra3
|
adhesion G protein-coupled receptor A3 |
| chr20_-_14545772 | 0.02 |
ENSRNOT00000001766
|
Bcr
|
BCR, RhoGEF and GTPase activating protein |
| chr8_+_33239139 | 0.02 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr5_-_79222687 | 0.02 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chr16_+_22250470 | 0.02 |
ENSRNOT00000015799
|
Lzts1
|
leucine zipper tumor suppressor 1 |
| chr4_-_11610518 | 0.02 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chrX_-_68562301 | 0.02 |
ENSRNOT00000076720
|
Ophn1
|
oligophrenin 1 |
| chr5_+_121952977 | 0.02 |
ENSRNOT00000008278
|
Pde4b
|
phosphodiesterase 4B |
| chr2_-_196377883 | 0.02 |
ENSRNOT00000028661
|
Gabpb2
|
GA binding protein transcription factor, beta subunit 2 |
| chr11_+_84396033 | 0.02 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chr8_-_48564722 | 0.02 |
ENSRNOT00000067902
|
Cbl
|
Cbl proto-oncogene |
| chr1_-_259484569 | 0.02 |
ENSRNOT00000021289
ENSRNOT00000021114 ENSRNOT00000021229 ENSRNOT00000021347 ENSRNOT00000021226 |
Sorbs1
|
sorbin and SH3 domain containing 1 |
| chr20_+_20105047 | 0.02 |
ENSRNOT00000082181
|
Ank3
|
ankyrin 3 |
| chr4_-_727691 | 0.02 |
ENSRNOT00000008497
|
Shh
|
sonic hedgehog |
| chr7_-_70577147 | 0.02 |
ENSRNOT00000008854
|
Mbd6
|
methyl-CpG binding domain protein 6 |
| chr2_-_125323454 | 0.02 |
ENSRNOT00000045837
|
Ankrd50
|
ankyrin repeat domain 50 |
| chr16_+_75966352 | 0.02 |
ENSRNOT00000022774
|
Angpt2
|
angiopoietin 2 |
| chr10_+_6975244 | 0.02 |
ENSRNOT00000046232
|
Usp7
|
ubiquitin specific peptidase 7 |
| chr10_+_46940965 | 0.02 |
ENSRNOT00000005366
|
Llgl1
|
LLGL1, scribble cell polarity complex component |
| chr17_+_31441630 | 0.02 |
ENSRNOT00000083705
ENSRNOT00000023582 |
Tubb2b
|
tubulin, beta 2B class IIb |
| chr18_-_72649915 | 0.02 |
ENSRNOT00000072205
|
AABR07032582.1
|
|
| chr18_-_69944632 | 0.02 |
ENSRNOT00000047271
|
Mapk4
|
mitogen-activated protein kinase 4 |
| chr9_-_38196273 | 0.02 |
ENSRNOT00000044452
|
Dst
|
dystonin |
| chr19_+_39543242 | 0.02 |
ENSRNOT00000018837
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr6_+_59757397 | 0.02 |
ENSRNOT00000047001
|
AABR07064000.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox3_Sox10
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.2 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0031632 | positive regulation of synaptic vesicle fusion to presynaptic membrane(GO:0031632) |
| 0.0 | 0.1 | GO:0072021 | ascending thin limb development(GO:0072021) metanephric ascending thin limb development(GO:0072218) |
| 0.0 | 0.0 | GO:1903210 | glomerular visceral epithelial cell apoptotic process(GO:1903210) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0099515 | actin filament-based transport(GO:0099515) |
| 0.0 | 0.0 | GO:0060769 | positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.0 | GO:0032900 | regulation of nucleoside transport(GO:0032242) negative regulation of neurotrophin production(GO:0032900) negative regulation of circadian sleep/wake cycle, non-REM sleep(GO:0042323) negative regulation of mucus secretion(GO:0070256) negative regulation of long term synaptic depression(GO:1900453) |
| 0.0 | 0.1 | GO:1904116 | response to vasopressin(GO:1904116) |
| 0.0 | 0.2 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.0 | 0.0 | GO:0031439 | positive regulation of mRNA cleavage(GO:0031439) positive regulation of endoribonuclease activity(GO:1902380) positive regulation of mRNA endonucleolytic cleavage involved in unfolded protein response(GO:1904722) |
| 0.0 | 0.0 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.0 | 0.0 | GO:1900095 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0003249 | left ventricular cardiac muscle tissue morphogenesis(GO:0003220) cell proliferation involved in heart valve morphogenesis(GO:0003249) regulation of cell proliferation involved in heart valve morphogenesis(GO:0003250) gonad morphogenesis(GO:0035262) nephrogenic mesenchyme morphogenesis(GO:0072134) |
| 0.0 | 0.0 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.0 | 0.0 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.0 | GO:0005943 | phosphatidylinositol 3-kinase complex, class IA(GO:0005943) |
| 0.0 | 0.0 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0098831 | presynaptic active zone cytoplasmic component(GO:0098831) |
| 0.0 | 0.0 | GO:1990907 | Scrib-APC-beta-catenin complex(GO:0034750) beta-catenin-TCF complex(GO:1990907) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005146 | leukemia inhibitory factor receptor binding(GO:0005146) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.1 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.0 | GO:0031249 | denatured protein binding(GO:0031249) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.0 | GO:0030350 | iron-responsive element binding(GO:0030350) |