Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
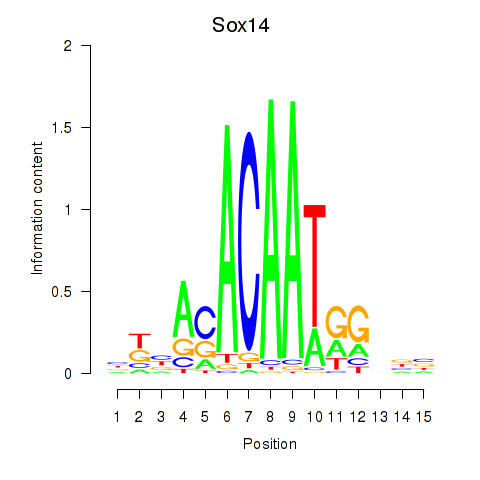
Results for Sox14
Z-value: 0.70
Transcription factors associated with Sox14
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox14
|
ENSRNOG00000022084 | SRY box 14 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox14 | rn6_v1_chr8_-_108109801_108109801 | 0.55 | 3.3e-01 | Click! |
Activity profile of Sox14 motif
Sorted Z-values of Sox14 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_88670430 | 1.38 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr3_-_152382529 | 0.64 |
ENSRNOT00000027076
|
Scand1
|
SCAN domain-containing 1 |
| chr1_-_190965115 | 0.42 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr1_+_81373340 | 0.32 |
ENSRNOT00000026814
|
Zfp428
|
zinc finger protein 428 |
| chr20_+_5049496 | 0.27 |
ENSRNOT00000088251
ENSRNOT00000001118 |
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr3_-_11417546 | 0.24 |
ENSRNOT00000018776
|
Lcn2
|
lipocalin 2 |
| chr10_-_96015499 | 0.24 |
ENSRNOT00000004383
|
Cacng4
|
calcium voltage-gated channel auxiliary subunit gamma 4 |
| chr10_+_55626741 | 0.24 |
ENSRNOT00000008492
|
Aurkb
|
aurora kinase B |
| chr12_-_2007516 | 0.23 |
ENSRNOT00000037564
|
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr1_-_216663720 | 0.22 |
ENSRNOT00000078944
ENSRNOT00000077409 |
Cdkn1c
|
cyclin-dependent kinase inhibitor 1C |
| chr7_+_116632506 | 0.22 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr16_-_6675746 | 0.22 |
ENSRNOT00000025858
|
Prkcd
|
protein kinase C, delta |
| chr17_+_70684340 | 0.22 |
ENSRNOT00000051067
|
Pfkfb3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 |
| chr7_-_54855557 | 0.21 |
ENSRNOT00000039475
|
Glipr1l1
|
GLI pathogenesis-related 1 like 1 |
| chr16_-_19399851 | 0.21 |
ENSRNOT00000089056
ENSRNOT00000021073 |
Tpm4
|
tropomyosin 4 |
| chr13_+_47602692 | 0.21 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr10_+_55627025 | 0.20 |
ENSRNOT00000091016
|
Aurkb
|
aurora kinase B |
| chr20_+_3422461 | 0.17 |
ENSRNOT00000084917
ENSRNOT00000079854 |
Tubb5
|
tubulin, beta 5 class I |
| chr10_-_90393317 | 0.17 |
ENSRNOT00000028563
|
Fam171a2
|
family with sequence similarity 171, member A2 |
| chr5_+_8459660 | 0.16 |
ENSRNOT00000007491
|
Cpa6
|
carboxypeptidase A6 |
| chr3_-_13435979 | 0.16 |
ENSRNOT00000029600
|
Pbx3
|
PBX homeobox 3 |
| chr5_-_173233188 | 0.16 |
ENSRNOT00000055343
|
Tmem88b
|
transmembrane protein 88B |
| chr1_+_81230612 | 0.14 |
ENSRNOT00000026489
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr9_+_98924134 | 0.14 |
ENSRNOT00000027597
|
Twist2
|
twist family bHLH transcription factor 2 |
| chr16_-_14300951 | 0.14 |
ENSRNOT00000017038
|
Rgr
|
retinal G protein coupled receptor |
| chr6_-_104290579 | 0.13 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr4_-_31730386 | 0.13 |
ENSRNOT00000013817
|
Slc25a13
|
solute carrier family 25 member 13 |
| chr1_-_47213749 | 0.13 |
ENSRNOT00000024656
|
Dynlt1
|
dynein light chain Tctex-type 1 |
| chr8_+_23113048 | 0.12 |
ENSRNOT00000029577
|
Cnn1
|
calponin 1 |
| chr4_-_113610243 | 0.12 |
ENSRNOT00000008813
|
Hk2
|
hexokinase 2 |
| chr2_+_159138758 | 0.12 |
ENSRNOT00000087942
|
AABR07011152.1
|
|
| chr20_+_13670066 | 0.12 |
ENSRNOT00000031400
|
LOC103694874
|
stromelysin-3 |
| chr8_+_65733400 | 0.12 |
ENSRNOT00000089126
|
Uaca
|
uveal autoantigen with coiled-coil domains and ankyrin repeats |
| chr1_-_59802401 | 0.11 |
ENSRNOT00000064917
|
Fpr2
|
formyl peptide receptor 2 |
| chr10_-_14085855 | 0.11 |
ENSRNOT00000018964
|
Rnf151
|
ring finger protein 151 |
| chr7_-_130350570 | 0.11 |
ENSRNOT00000055805
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr10_-_34961608 | 0.11 |
ENSRNOT00000033056
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr9_+_27546528 | 0.11 |
ENSRNOT00000051403
|
Khdc1
|
KH homology domain containing 1 |
| chr17_+_31441630 | 0.10 |
ENSRNOT00000083705
ENSRNOT00000023582 |
Tubb2b
|
tubulin, beta 2B class IIb |
| chr16_-_47535358 | 0.10 |
ENSRNOT00000040731
|
Cldn22
|
claudin 22 |
| chr13_-_36174908 | 0.10 |
ENSRNOT00000072295
|
Dbi
|
diazepam binding inhibitor, acyl-CoA binding protein |
| chr5_-_153625869 | 0.10 |
ENSRNOT00000024464
|
Clic4
|
chloride intracellular channel 4 |
| chr17_-_2705123 | 0.10 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr16_-_20890949 | 0.10 |
ENSRNOT00000081977
|
Homer3
|
homer scaffolding protein 3 |
| chr2_-_104133985 | 0.10 |
ENSRNOT00000088167
ENSRNOT00000067725 |
Pde7a
|
phosphodiesterase 7A |
| chr13_-_72789841 | 0.10 |
ENSRNOT00000004694
|
Mr1
|
major histocompatibility complex, class I-related |
| chrM_+_7758 | 0.09 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr17_+_14469488 | 0.09 |
ENSRNOT00000060670
|
AABR07027111.1
|
|
| chr4_+_30313102 | 0.09 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr1_+_221735517 | 0.09 |
ENSRNOT00000028628
ENSRNOT00000044866 |
Sf1
|
splicing factor 1 |
| chr10_+_7041510 | 0.09 |
ENSRNOT00000003514
|
Carhsp1
|
calcium regulated heat stable protein 1 |
| chr7_-_11513581 | 0.09 |
ENSRNOT00000027045
|
Thop1
|
thimet oligopeptidase 1 |
| chr6_+_104437159 | 0.09 |
ENSRNOT00000076485
|
Susd6
|
sushi domain containing 6 |
| chr4_-_727691 | 0.09 |
ENSRNOT00000008497
|
Shh
|
sonic hedgehog |
| chr2_+_38230757 | 0.09 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr20_+_31234493 | 0.08 |
ENSRNOT00000000675
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr14_-_19072677 | 0.08 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr7_-_144959746 | 0.08 |
ENSRNOT00000064261
|
Zfp385a
|
zinc finger protein 385A |
| chr3_+_112242270 | 0.08 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr20_+_3553455 | 0.08 |
ENSRNOT00000090080
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chrX_+_110789269 | 0.08 |
ENSRNOT00000086014
|
Rnf128
|
ring finger protein 128, E3 ubiquitin protein ligase |
| chr20_-_4380600 | 0.08 |
ENSRNOT00000068712
|
Egfl8
|
EGF-like-domain, multiple 8 |
| chr17_-_43627629 | 0.08 |
ENSRNOT00000022965
|
Hist1h2af
|
histone cluster 1, H2af |
| chr10_-_93675991 | 0.08 |
ENSRNOT00000090662
|
March10
|
membrane associated ring-CH-type finger 10 |
| chr10_+_105498728 | 0.08 |
ENSRNOT00000032163
|
Sphk1
|
sphingosine kinase 1 |
| chr12_+_2046472 | 0.08 |
ENSRNOT00000001289
|
Zfp358
|
zinc finger protein 358 |
| chr8_+_97291580 | 0.07 |
ENSRNOT00000018794
|
Rasgrf1
|
RAS protein-specific guanine nucleotide-releasing factor 1 |
| chr1_-_98501249 | 0.07 |
ENSRNOT00000023858
|
Lim2
|
lens intrinsic membrane protein 2 |
| chr1_+_21525421 | 0.07 |
ENSRNOT00000017911
|
Arg1
|
arginase 1 |
| chr19_+_61677542 | 0.07 |
ENSRNOT00000014785
|
Itgb1
|
integrin subunit beta 1 |
| chr4_+_64088900 | 0.07 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr9_+_46657922 | 0.07 |
ENSRNOT00000019180
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr14_-_6148830 | 0.07 |
ENSRNOT00000075091
ENSRNOT00000092683 |
LOC100366030
|
rCG37858-like |
| chr12_-_52644260 | 0.07 |
ENSRNOT00000073986
|
Plcxd1
|
phosphatidylinositol-specific phospholipase C, X domain containing 1 |
| chr14_+_44413636 | 0.07 |
ENSRNOT00000003552
|
Smim14
|
small integral membrane protein 14 |
| chr10_-_56267213 | 0.07 |
ENSRNOT00000017201
|
Mpdu1
|
mannose-P-dolichol utilization defect 1 |
| chrX_+_105402867 | 0.07 |
ENSRNOT00000057222
|
Rpl36a
|
ribosomal protein L36a |
| chr1_-_163328591 | 0.07 |
ENSRNOT00000034843
|
Tsku
|
tsukushi, small leucine rich proteoglycan |
| chr5_+_152681101 | 0.07 |
ENSRNOT00000076052
ENSRNOT00000022574 |
Stmn1
|
stathmin 1 |
| chr4_-_157263890 | 0.07 |
ENSRNOT00000065416
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr4_+_115388839 | 0.07 |
ENSRNOT00000018128
|
Vax2
|
ventral anterior homeobox 2 |
| chr3_-_119075606 | 0.06 |
ENSRNOT00000041569
ENSRNOT00000086724 |
Hdc
|
histidine decarboxylase |
| chr6_+_55374984 | 0.06 |
ENSRNOT00000091682
|
Agr3
|
anterior gradient 3, protein disulphide isomerase family member |
| chr1_+_226234074 | 0.06 |
ENSRNOT00000027834
|
Fads1
|
fatty acid desaturase 1 |
| chr9_+_46657575 | 0.06 |
ENSRNOT00000083322
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr6_+_137164535 | 0.06 |
ENSRNOT00000018225
|
Inf2
|
inverted formin, FH2 and WH2 domain containing |
| chr4_-_170841187 | 0.06 |
ENSRNOT00000007485
|
Art4
|
ADP-ribosyltransferase 4 |
| chr7_+_116355911 | 0.06 |
ENSRNOT00000076182
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr16_+_6035926 | 0.06 |
ENSRNOT00000020284
|
Selenok
|
selenoprotein K |
| chr2_-_257376756 | 0.06 |
ENSRNOT00000065811
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr6_+_111296417 | 0.06 |
ENSRNOT00000075425
|
Ahsa1
|
activator of Hsp90 ATPase activity 1 |
| chr7_-_12399910 | 0.06 |
ENSRNOT00000021677
|
RGD1562114
|
RGD1562114 |
| chr4_+_155321553 | 0.06 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr5_+_129257429 | 0.06 |
ENSRNOT00000072396
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr10_-_103340922 | 0.06 |
ENSRNOT00000004191
|
Btbd17
|
BTB domain containing 17 |
| chr8_-_111965889 | 0.06 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr1_-_190914610 | 0.05 |
ENSRNOT00000023189
|
Cdr2
|
cerebellar degeneration-related protein 2 |
| chr6_-_1942972 | 0.05 |
ENSRNOT00000048711
|
Cdc42ep3
|
CDC42 effector protein 3 |
| chr9_+_17784468 | 0.05 |
ENSRNOT00000026831
|
Slc29a1
|
solute carrier family 29 member 1 |
| chr4_-_77706994 | 0.05 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr1_-_198869009 | 0.05 |
ENSRNOT00000024858
|
Zfp688
|
zinc finger protein 688 |
| chr8_+_116325286 | 0.05 |
ENSRNOT00000017461
|
Hyal2
|
hyaluronoglucosaminidase 2 |
| chr14_+_77380262 | 0.05 |
ENSRNOT00000008030
|
Nsg1
|
neuron specific gene family member 1 |
| chr8_-_33661049 | 0.05 |
ENSRNOT00000068037
|
Fli1
|
Fli-1 proto-oncogene, ETS transcription factor |
| chr10_-_86393141 | 0.05 |
ENSRNOT00000009485
|
Mien1
|
migration and invasion enhancer 1 |
| chr3_+_176691329 | 0.05 |
ENSRNOT00000017319
|
Ppdpf
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr9_-_113504605 | 0.05 |
ENSRNOT00000067974
|
Rab31
|
RAB31, member RAS oncogene family |
| chr14_-_85350948 | 0.05 |
ENSRNOT00000083464
|
LOC100912481
|
RNA-binding protein EWS-like |
| chr6_+_135890931 | 0.05 |
ENSRNOT00000042973
|
Tnfaip2
|
TNF alpha induced protein 2 |
| chr4_-_7228675 | 0.05 |
ENSRNOT00000064386
|
Agap3
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chrX_+_48196916 | 0.05 |
ENSRNOT00000045976
|
Da2-19
|
uncharacterized LOC302729 |
| chr4_+_24612205 | 0.05 |
ENSRNOT00000057382
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chrX_+_28593405 | 0.05 |
ENSRNOT00000071708
|
Tmsb4x
|
thymosin beta 4, X-linked |
| chr19_+_56024903 | 0.05 |
ENSRNOT00000021607
|
Cdk10
|
cyclin-dependent kinase 10 |
| chr20_+_42966140 | 0.05 |
ENSRNOT00000000707
|
Marcks
|
myristoylated alanine rich protein kinase C substrate |
| chr6_-_123894826 | 0.05 |
ENSRNOT00000082444
|
Efcab11
|
EF-hand calcium binding domain 11 |
| chr1_-_201906286 | 0.05 |
ENSRNOT00000064511
ENSRNOT00000036625 |
RGD1560958
|
similar to RIKEN cDNA 1700063I17 |
| chr9_-_80167033 | 0.05 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr8_-_13606040 | 0.05 |
ENSRNOT00000013577
ENSRNOT00000083395 |
Panx1
|
Pannexin 1 |
| chr8_-_19917396 | 0.05 |
ENSRNOT00000032963
|
Olr1158
|
olfactory receptor 1158 |
| chr3_-_72219246 | 0.04 |
ENSRNOT00000009903
|
Smtnl1
|
smoothelin-like 1 |
| chr6_-_10891146 | 0.04 |
ENSRNOT00000020328
|
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr15_+_57891680 | 0.04 |
ENSRNOT00000001383
|
Tpt1
|
tumor protein, translationally-controlled 1 |
| chr17_-_44840131 | 0.04 |
ENSRNOT00000083417
|
Hist1h3b
|
histone cluster 1, H3b |
| chr18_+_30527705 | 0.04 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr18_-_37776453 | 0.04 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr4_+_78694447 | 0.04 |
ENSRNOT00000011945
|
Gpnmb
|
glycoprotein nmb |
| chr16_-_74122889 | 0.04 |
ENSRNOT00000025763
|
Plat
|
plasminogen activator, tissue type |
| chrX_-_152367861 | 0.04 |
ENSRNOT00000089414
|
LOC103690371
|
melanoma-associated antigen 10-like |
| chr1_-_61686944 | 0.04 |
ENSRNOT00000059638
|
Vom1r23
|
vomeronasal 1 receptor 23 |
| chr20_-_4368693 | 0.04 |
ENSRNOT00000000501
|
Rnf5
|
ring finger protein 5 |
| chr1_-_67065797 | 0.04 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr7_+_116356324 | 0.04 |
ENSRNOT00000076407
ENSRNOT00000077022 ENSRNOT00000075873 |
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr18_+_30509393 | 0.04 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr7_+_14643704 | 0.04 |
ENSRNOT00000044642
|
AABR07055875.1
|
|
| chr18_+_55466373 | 0.04 |
ENSRNOT00000074629
|
LOC102555392
|
interferon-inducible GTPase 1-like |
| chr7_+_11521998 | 0.04 |
ENSRNOT00000026960
|
Sgta
|
small glutamine rich tetratricopeptide repeat containing alpha |
| chr6_+_8669722 | 0.04 |
ENSRNOT00000048550
|
Camkmt
|
calmodulin-lysine N-methyltransferase |
| chr10_+_78168715 | 0.04 |
ENSRNOT00000072294
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr1_+_78546012 | 0.04 |
ENSRNOT00000058172
|
Gng5
|
G protein subunit gamma 5 |
| chr11_+_67188630 | 0.04 |
ENSRNOT00000003092
|
Casr
|
calcium-sensing receptor |
| chr6_-_139142218 | 0.04 |
ENSRNOT00000006975
|
Ighg
|
Immunoglobulin heavy chain (gamma polypeptide) |
| chrX_-_156456035 | 0.04 |
ENSRNOT00000080990
|
Emd
|
emerin |
| chr2_+_60180215 | 0.04 |
ENSRNOT00000084624
|
Prlr
|
prolactin receptor |
| chr1_-_62681471 | 0.03 |
ENSRNOT00000042466
|
Vom2r15
|
vomeronasal 2 receptor, 15 |
| chr3_-_151658417 | 0.03 |
ENSRNOT00000047246
|
LOC100910990
|
copine-1-like |
| chr10_-_58834538 | 0.03 |
ENSRNOT00000020043
|
Slc13a5
|
solute carrier family 13 member 5 |
| chr4_+_161696903 | 0.03 |
ENSRNOT00000055889
|
LOC100362909
|
rCG30099-like |
| chr2_+_189430041 | 0.03 |
ENSRNOT00000023567
ENSRNOT00000023605 |
Tpm3
|
tropomyosin 3 |
| chr1_-_219438779 | 0.03 |
ENSRNOT00000029237
|
Tbc1d10c
|
TBC1 domain family, member 10C |
| chr12_-_19114399 | 0.03 |
ENSRNOT00000073099
|
Cyp3a9
|
cytochrome P450, family 3, subfamily a, polypeptide 9 |
| chr12_+_7454884 | 0.03 |
ENSRNOT00000077328
|
LOC100910196
|
katanin p60 ATPase-containing subunit A-like 1-like |
| chr5_+_129052043 | 0.03 |
ENSRNOT00000013459
|
LOC108348114
|
tetratricopeptide repeat protein 39A |
| chr5_-_158549496 | 0.03 |
ENSRNOT00000074336
|
Igsf21
|
immunoglobin superfamily, member 21 |
| chr10_-_109979712 | 0.03 |
ENSRNOT00000086042
|
Dus1l
|
dihydrouridine synthase 1-like |
| chr14_-_85350786 | 0.03 |
ENSRNOT00000012634
|
LOC100912481
|
RNA-binding protein EWS-like |
| chr2_-_41871858 | 0.03 |
ENSRNOT00000039720
|
Gapt
|
Grb2-binding adaptor protein, transmembrane |
| chr15_+_52220578 | 0.03 |
ENSRNOT00000015104
|
Lgi3
|
leucine-rich repeat LGI family, member 3 |
| chr18_+_29993361 | 0.03 |
ENSRNOT00000075810
|
Pcdha4
|
protocadherin alpha 4 |
| chr3_-_3747440 | 0.03 |
ENSRNOT00000036178
|
Ccdc187
|
coiled-coil domain containing 187 |
| chr1_-_60828260 | 0.03 |
ENSRNOT00000059671
|
Vom1r-ps112
|
vomeronasal 1 receptor pseudogene 112 |
| chr5_-_173484986 | 0.03 |
ENSRNOT00000064207
ENSRNOT00000055319 |
Ttll10
|
tubulin tyrosine ligase like 10 |
| chr7_-_114380613 | 0.03 |
ENSRNOT00000011898
|
Ago2
|
argonaute 2, RISC catalytic component |
| chr19_-_17115113 | 0.03 |
ENSRNOT00000015718
|
Fto
|
FTO, alpha-ketoglutarate dependent dioxygenase |
| chr4_-_178168690 | 0.03 |
ENSRNOT00000020729
|
Sox5
|
SRY box 5 |
| chr10_+_92337879 | 0.03 |
ENSRNOT00000042984
|
Mapt
|
microtubule-associated protein tau |
| chr14_-_80958478 | 0.03 |
ENSRNOT00000035385
|
Dok7
|
docking protein 7 |
| chr7_+_116355698 | 0.03 |
ENSRNOT00000076693
|
Ly6e
|
lymphocyte antigen 6 complex, locus E |
| chr3_-_146690375 | 0.03 |
ENSRNOT00000010641
|
Abhd12
|
abhydrolase domain containing 12 |
| chrX_+_62727755 | 0.03 |
ENSRNOT00000077055
|
Pcyt1b
|
phosphate cytidylyltransferase 1, choline, beta |
| chr16_-_22561496 | 0.03 |
ENSRNOT00000016543
|
Lpl
|
lipoprotein lipase |
| chr6_-_38453797 | 0.03 |
ENSRNOT00000009100
|
Ddx1
|
DEAD-box helicase 1 |
| chr17_+_2690062 | 0.03 |
ENSRNOT00000024937
|
Olr1653
|
olfactory receptor 1653 |
| chrX_-_70835089 | 0.03 |
ENSRNOT00000076351
|
Tex11
|
testis expressed 11 |
| chr11_+_46793777 | 0.03 |
ENSRNOT00000002232
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr1_+_63263722 | 0.03 |
ENSRNOT00000088203
|
LOC100911184
|
vomeronasal type-1 receptor 4-like |
| chr13_-_91776397 | 0.03 |
ENSRNOT00000073147
|
Mptx1
|
mucosal pentraxin 1 |
| chr6_+_132510757 | 0.03 |
ENSRNOT00000080230
|
Evl
|
Enah/Vasp-like |
| chr8_+_42511520 | 0.03 |
ENSRNOT00000042974
|
Olr1259
|
olfactory receptor 1259 |
| chr4_+_56591715 | 0.03 |
ENSRNOT00000041207
|
Fam71f1
|
family with sequence similarity 71, member F1 |
| chr7_+_26375866 | 0.03 |
ENSRNOT00000059639
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr8_+_22966561 | 0.03 |
ENSRNOT00000016819
|
Swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr10_-_37055282 | 0.03 |
ENSRNOT00000086193
ENSRNOT00000065584 |
Hnrnpab
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr12_+_38160464 | 0.03 |
ENSRNOT00000032249
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chr3_-_119135391 | 0.03 |
ENSRNOT00000045443
|
Gabpb1
|
GA binding protein transcription factor, beta subunit 1 |
| chr19_+_24329544 | 0.03 |
ENSRNOT00000080934
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr12_+_10255416 | 0.02 |
ENSRNOT00000092720
|
Gpr12
|
G protein-coupled receptor 12 |
| chr8_-_65587427 | 0.02 |
ENSRNOT00000016491
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr5_+_145188323 | 0.02 |
ENSRNOT00000038888
|
Tmem35b
|
transmembrane protein 35B |
| chr10_-_18506337 | 0.02 |
ENSRNOT00000043036
|
Gabrp
|
gamma-aminobutyric acid type A receptor pi subunit |
| chr19_-_49623758 | 0.02 |
ENSRNOT00000071130
|
Pkd1l2
|
polycystic kidney disease 1-like 2 |
| chr8_-_65587658 | 0.02 |
ENSRNOT00000091982
|
Lrrc49
|
leucine rich repeat containing 49 |
| chr9_-_61810417 | 0.02 |
ENSRNOT00000020910
|
Rftn2
|
raftlin family member 2 |
| chr1_+_190964885 | 0.02 |
ENSRNOT00000039186
|
Mettl9
|
methyltransferase like 9 |
| chr2_-_100249811 | 0.02 |
ENSRNOT00000086760
|
Hnf4g
|
hepatocyte nuclear factor 4, gamma |
| chr1_+_68239314 | 0.02 |
ENSRNOT00000070823
|
Vom1r109
|
vomeronasal 1 receptor 109 |
| chr14_+_108007724 | 0.02 |
ENSRNOT00000014062
|
Xpo1
|
exportin 1 |
| chr4_-_138830117 | 0.02 |
ENSRNOT00000008403
|
Il5ra
|
interleukin 5 receptor subunit alpha |
| chr6_-_43493816 | 0.02 |
ENSRNOT00000077786
|
Ywhaq
|
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta |
| chr10_-_34961349 | 0.02 |
ENSRNOT00000004885
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox14
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.4 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.1 | 0.4 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.2 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.1 | 0.2 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.1 | 0.2 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.2 | GO:1902746 | regulation of lens fiber cell differentiation(GO:1902746) |
| 0.0 | 0.1 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.0 | 0.1 | GO:0019060 | intracellular transport of viral protein in host cell(GO:0019060) symbiont intracellular protein transport in host(GO:0030581) intracellular protein transport in other organism involved in symbiotic interaction(GO:0051708) |
| 0.0 | 0.1 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.1 | GO:0016539 | intein-mediated protein splicing(GO:0016539) protein splicing(GO:0030908) positive regulation of epithelial cell proliferation involved in prostate gland development(GO:0060769) specification of axis polarity(GO:0065001) negative regulation of cholesterol efflux(GO:0090370) negative regulation of dopaminergic neuron differentiation(GO:1904339) positive regulation of mesenchymal cell proliferation involved in ureter development(GO:2000729) |
| 0.0 | 0.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.3 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.1 | GO:0036151 | phosphatidylcholine acyl-chain remodeling(GO:0036151) |
| 0.0 | 0.1 | GO:0007161 | calcium-independent cell-matrix adhesion(GO:0007161) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.1 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.2 | GO:1990001 | inhibition of cysteine-type endopeptidase activity involved in apoptotic process(GO:1990001) |
| 0.0 | 0.1 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.1 | GO:0070494 | regulation of thrombin receptor signaling pathway(GO:0070494) negative regulation of thrombin receptor signaling pathway(GO:0070495) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) histidine catabolic process(GO:0006548) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:2000383 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
| 0.0 | 0.1 | GO:0050862 | positive regulation of T cell receptor signaling pathway(GO:0050862) |
| 0.0 | 0.0 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.0 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.1 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.0 | GO:1904204 | regulation of skeletal muscle hypertrophy(GO:1904204) |
| 0.0 | 0.0 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.0 | GO:0015744 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.0 | GO:0060279 | positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.1 | GO:0048242 | epinephrine secretion(GO:0048242) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.1 | GO:0034667 | integrin alpha2-beta1 complex(GO:0034666) integrin alpha3-beta1 complex(GO:0034667) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.3 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.1 | GO:0043293 | apoptosome(GO:0043293) |
| 0.0 | 0.0 | GO:0031680 | G-protein beta/gamma-subunit complex(GO:0031680) |
| 0.0 | 0.2 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.0 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 1.3 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0098845 | postsynaptic endosome(GO:0098845) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.2 | GO:0070976 | TIR domain binding(GO:0070976) |
| 0.1 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004982 | N-formyl peptide receptor activity(GO:0004982) |
| 0.0 | 0.2 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.4 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.1 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0033906 | hyaluronoglucuronidase activity(GO:0033906) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.2 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.1 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.0 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.1 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.0 | REACTOME NEP NS2 INTERACTS WITH THE CELLULAR EXPORT MACHINERY | Genes involved in NEP/NS2 Interacts with the Cellular Export Machinery |