Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
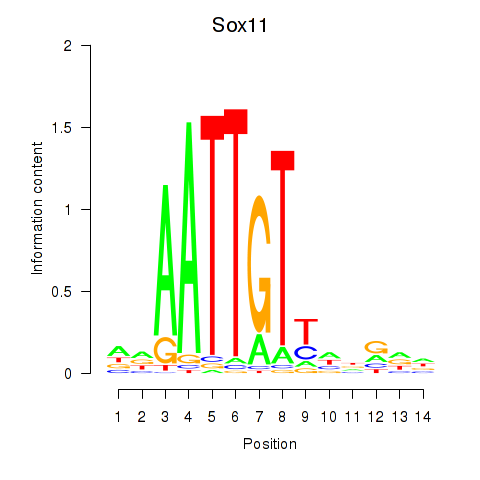
Results for Sox11
Z-value: 0.37
Transcription factors associated with Sox11
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Sox11
|
ENSRNOG00000030034 | SRY box 11 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Sox11 | rn6_v1_chr6_-_46631983_46631983 | 0.48 | 4.2e-01 | Click! |
Activity profile of Sox11 motif
Sorted Z-values of Sox11 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_10335499 | 0.19 |
ENSRNOT00000071567
|
Wasf3
|
WAS protein family, member 3 |
| chr1_-_277902279 | 0.14 |
ENSRNOT00000078416
|
Ablim1
|
actin-binding LIM protein 1 |
| chr18_+_14471213 | 0.11 |
ENSRNOT00000045224
|
Dtna
|
dystrobrevin, alpha |
| chr10_-_58771908 | 0.09 |
ENSRNOT00000018808
|
RGD1304728
|
similar to 4933427D14Rik protein |
| chr10_-_35800120 | 0.08 |
ENSRNOT00000004361
|
Maml1
|
mastermind-like transcriptional coactivator 1 |
| chr6_-_106971250 | 0.08 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr1_-_235405831 | 0.08 |
ENSRNOT00000071578
|
AABR07006458.1
|
|
| chr16_+_60925093 | 0.08 |
ENSRNOT00000015813
|
Tnks
|
tankyrase |
| chr6_-_76552559 | 0.08 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr16_+_50152008 | 0.07 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr19_-_17494516 | 0.07 |
ENSRNOT00000078786
ENSRNOT00000076138 |
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr4_-_11497531 | 0.07 |
ENSRNOT00000078799
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr16_-_74020437 | 0.06 |
ENSRNOT00000037389
|
Kat6a
|
lysine acetyltransferase 6A |
| chr2_+_185846232 | 0.06 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr2_+_4989295 | 0.06 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
| chr8_+_109455786 | 0.06 |
ENSRNOT00000039593
|
Msl2
|
male-specific lethal 2 homolog (Drosophila) |
| chr2_+_56052643 | 0.06 |
ENSRNOT00000029565
|
Rictor
|
RPTOR independent companion of MTOR, complex 2 |
| chr5_+_2278357 | 0.06 |
ENSRNOT00000066590
|
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr10_-_49149479 | 0.06 |
ENSRNOT00000004291
|
Zfp287
|
zinc finger protein 287 |
| chr1_-_124038966 | 0.05 |
ENSRNOT00000082487
|
Chrna7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chrX_+_65040934 | 0.05 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr2_+_114386019 | 0.05 |
ENSRNOT00000082148
|
AABR07009834.1
|
|
| chr14_+_8080565 | 0.05 |
ENSRNOT00000092395
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr5_-_160070916 | 0.05 |
ENSRNOT00000067517
ENSRNOT00000055791 |
Spen
|
spen family transcriptional repressor |
| chr2_+_189655702 | 0.05 |
ENSRNOT00000021502
|
Gatad2b
|
GATA zinc finger domain containing 2B |
| chr3_-_66417741 | 0.05 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chrX_+_68752597 | 0.04 |
ENSRNOT00000077039
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr14_+_23507628 | 0.04 |
ENSRNOT00000037509
|
Uba6
|
ubiquitin-like modifier activating enzyme 6 |
| chr11_+_66316606 | 0.04 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr7_-_122329443 | 0.04 |
ENSRNOT00000032232
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr2_-_54823917 | 0.04 |
ENSRNOT00000039057
ENSRNOT00000079333 |
Card6
|
caspase recruitment domain family, member 6 |
| chr1_-_124039196 | 0.04 |
ENSRNOT00000051883
|
Chrna7
|
cholinergic receptor nicotinic alpha 7 subunit |
| chr5_+_64566804 | 0.04 |
ENSRNOT00000073192
|
LOC103690035
|
TGF-beta receptor type-1-like |
| chr13_+_89805962 | 0.04 |
ENSRNOT00000074035
|
Tstd1
|
thiosulfate sulfurtransferase like domain containing 1 |
| chr4_-_16669368 | 0.03 |
ENSRNOT00000007608
|
Pclo
|
piccolo (presynaptic cytomatrix protein) |
| chr3_-_152179193 | 0.03 |
ENSRNOT00000026700
|
Rbm12
|
RNA binding motif protein 12 |
| chr10_-_52290657 | 0.03 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr2_+_22950018 | 0.03 |
ENSRNOT00000071804
|
Homer1
|
homer scaffolding protein 1 |
| chr5_+_171274544 | 0.03 |
ENSRNOT00000077307
|
Cep104
|
centrosomal protein 104 |
| chr5_+_35991068 | 0.03 |
ENSRNOT00000061139
|
Pnisr
|
PNN interacting serine and arginine rich protein |
| chr19_+_39543242 | 0.03 |
ENSRNOT00000018837
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr5_-_117754571 | 0.03 |
ENSRNOT00000011405
|
Dock7
|
dedicator of cytokinesis 7 |
| chr13_+_57243877 | 0.03 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr11_-_1983513 | 0.03 |
ENSRNOT00000000907
|
Htr1f
|
5-hydroxytryptamine receptor 1F |
| chr2_+_27005863 | 0.03 |
ENSRNOT00000080864
|
Poc5
|
POC5 centriolar protein |
| chr13_-_82295123 | 0.03 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr15_-_108120279 | 0.03 |
ENSRNOT00000090572
|
Dock9
|
dedicator of cytokinesis 9 |
| chr6_-_78817641 | 0.03 |
ENSRNOT00000072117
|
AABR07064392.1
|
|
| chr17_-_77527894 | 0.03 |
ENSRNOT00000032173
|
Bend7
|
BEN domain containing 7 |
| chr3_-_100364400 | 0.03 |
ENSRNOT00000006670
|
Mettl15
|
methyltransferase like 15 |
| chr17_+_82065937 | 0.03 |
ENSRNOT00000051349
|
Arl5b
|
ADP-ribosylation factor like GTPase 5B |
| chrX_+_10042893 | 0.03 |
ENSRNOT00000084218
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr20_-_6257604 | 0.02 |
ENSRNOT00000092489
|
Stk38
|
serine/threonine kinase 38 |
| chr6_+_137924568 | 0.02 |
ENSRNOT00000046456
|
Mta1
|
metastasis associated 1 |
| chr3_+_80468154 | 0.02 |
ENSRNOT00000021837
|
Ckap5
|
cytoskeleton associated protein 5 |
| chr2_+_18937458 | 0.02 |
ENSRNOT00000022377
|
Tmem167a
|
transmembrane protein 167A |
| chrX_+_9956370 | 0.02 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr8_-_14880644 | 0.02 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr3_-_57913999 | 0.02 |
ENSRNOT00000090426
|
Slc25a12
|
solute carrier family 25 member 12 |
| chr17_-_47653989 | 0.02 |
ENSRNOT00000072833
|
LOC498759
|
LRRGT00094 |
| chr15_-_93748742 | 0.02 |
ENSRNOT00000093370
|
Mycbp2
|
MYC binding protein 2, E3 ubiquitin protein ligase |
| chr1_+_171793782 | 0.01 |
ENSRNOT00000081351
|
Olfml1
|
olfactomedin-like 1 |
| chr14_-_45165207 | 0.01 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr14_+_8080275 | 0.01 |
ENSRNOT00000065965
ENSRNOT00000092542 |
Mapk10
|
mitogen activated protein kinase 10 |
| chr2_+_206454208 | 0.01 |
ENSRNOT00000026863
|
Phtf1
|
putative homeodomain transcription factor 1 |
| chr6_-_92398525 | 0.01 |
ENSRNOT00000007314
|
Sav1
|
salvador family WW domain containing protein 1 |
| chr20_-_45053640 | 0.01 |
ENSRNOT00000072256
|
RGD1561777
|
similar to Na+ dependent glucose transporter 1 |
| chr7_-_119689938 | 0.01 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr5_-_144996431 | 0.01 |
ENSRNOT00000016992
|
Zmym4
|
zinc finger MYM-type containing 4 |
| chr17_-_32015071 | 0.01 |
ENSRNOT00000070876
|
LOC100911107
|
leukocyte elastase inhibitor A-like |
| chr12_-_12546111 | 0.01 |
ENSRNOT00000046381
|
Lmtk2
|
lemur tyrosine kinase 2 |
| chrM_+_11736 | 0.01 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr7_+_65616177 | 0.01 |
ENSRNOT00000006566
|
Gns
|
glucosamine (N-acetyl)-6-sulfatase |
| chr18_-_26656879 | 0.01 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr2_-_183210799 | 0.01 |
ENSRNOT00000085382
|
Trim2
|
tripartite motif-containing 2 |
| chr4_+_22082194 | 0.01 |
ENSRNOT00000091799
|
Crot
|
carnitine O-octanoyltransferase |
| chr17_-_14627937 | 0.00 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr6_+_100337226 | 0.00 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chr4_-_164051812 | 0.00 |
ENSRNOT00000085719
|
AABR07062183.1
|
|
| chr10_+_93520132 | 0.00 |
ENSRNOT00000055137
|
Mrc2
|
mannose receptor, C type 2 |
| chrX_+_65040775 | 0.00 |
ENSRNOT00000081354
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
Network of associatons between targets according to the STRING database.
First level regulatory network of Sox11
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) regulation of telomeric DNA binding(GO:1904742) |
| 0.0 | 0.1 | GO:0060926 | cardiac pacemaker cell development(GO:0060926) |
| 0.0 | 0.1 | GO:0098964 | dendritic transport of ribonucleoprotein complex(GO:0098961) dendritic transport of messenger ribonucleoprotein complex(GO:0098963) anterograde dendritic transport of messenger ribonucleoprotein complex(GO:0098964) |
| 0.0 | 0.1 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.2 | GO:0031643 | positive regulation of myelination(GO:0031643) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0044317 | rod spherule(GO:0044317) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.0 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |