Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
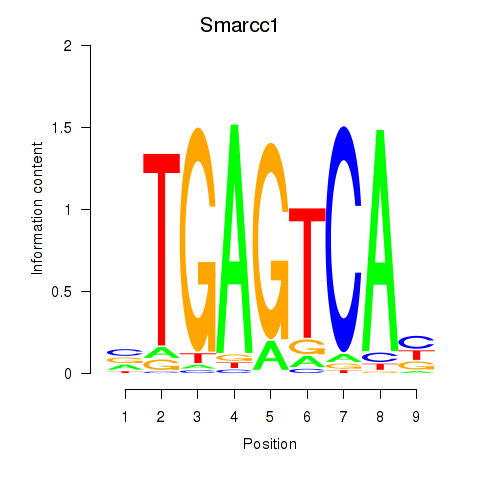
Results for Smarcc1_Fosl1
Z-value: 0.72
Transcription factors associated with Smarcc1_Fosl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smarcc1
|
ENSRNOG00000020804 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
|
Fosl1
|
ENSRNOG00000020552 | FOS like 1, AP-1 transcription factor subunit |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smarcc1 | rn6_v1_chr8_+_118206698_118206703 | -0.26 | 6.7e-01 | Click! |
| Fosl1 | rn6_v1_chr1_+_220826560_220826560 | 0.09 | 8.9e-01 | Click! |
Activity profile of Smarcc1_Fosl1 motif
Sorted Z-values of Smarcc1_Fosl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_145870912 | 0.59 |
ENSRNOT00000016289
|
Il16
|
interleukin 16 |
| chr12_-_48365784 | 0.56 |
ENSRNOT00000077317
|
Dao
|
D-amino-acid oxidase |
| chr20_+_5067330 | 0.55 |
ENSRNOT00000037482
|
Ly6g6e
|
lymphocyte antigen 6 complex, locus G6E |
| chr4_+_86275717 | 0.53 |
ENSRNOT00000016414
|
Ppp1r17
|
protein phosphatase 1, regulatory subunit 17 |
| chr2_+_190003223 | 0.50 |
ENSRNOT00000015712
|
S100a5
|
S100 calcium binding protein A5 |
| chr17_-_2705123 | 0.30 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr3_+_161121697 | 0.29 |
ENSRNOT00000036020
|
Wfdc10a
|
WAP four-disulfide core domain 10A |
| chr1_+_12006660 | 0.25 |
ENSRNOT00000093439
|
Mcc
|
colorectal mutant cancer protein |
| chr1_+_218466289 | 0.24 |
ENSRNOT00000017948
|
Mrgprf
|
MAS related GPR family member F |
| chr9_+_15166118 | 0.22 |
ENSRNOT00000020432
|
Mdfi
|
MyoD family inhibitor |
| chr3_-_64554953 | 0.21 |
ENSRNOT00000067452
|
LOC102553814
|
serine/arginine repetitive matrix protein 1-like |
| chr1_+_215666628 | 0.21 |
ENSRNOT00000040598
ENSRNOT00000066135 ENSRNOT00000051425 ENSRNOT00000080339 ENSRNOT00000066896 ENSRNOT00000063918 |
Tnnt3
|
troponin T3, fast skeletal type |
| chr18_-_27206777 | 0.21 |
ENSRNOT00000090569
|
AABR07031691.1
|
|
| chr19_-_37250919 | 0.21 |
ENSRNOT00000021015
|
Exoc3l1
|
exocyst complex component 3-like 1 |
| chr9_+_25410669 | 0.21 |
ENSRNOT00000030912
ENSRNOT00000090920 |
Tfap2b
|
transcription factor AP-2 beta |
| chr20_+_3230052 | 0.20 |
ENSRNOT00000078454
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr1_-_91074294 | 0.19 |
ENSRNOT00000075236
|
LOC687508
|
similar to Cytochrome c oxidase polypeptide VIIa-heart, mitochondrial precursor (Cytochrome c oxidase subunit VIIa-H) (COX VIIa-M) |
| chr7_+_59200918 | 0.19 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr4_+_181243338 | 0.19 |
ENSRNOT00000034051
|
Smco2
|
single-pass membrane protein with coiled-coil domains 2 |
| chr1_-_170318935 | 0.18 |
ENSRNOT00000024119
|
Prkcdbp
|
protein kinase C, delta binding protein |
| chr20_+_40618623 | 0.18 |
ENSRNOT00000082043
|
Pkib
|
cAMP-dependent protein kinase inhibitor beta |
| chr15_-_34444244 | 0.18 |
ENSRNOT00000027612
|
Cideb
|
cell death-inducing DFFA-like effector b |
| chr1_-_124803363 | 0.18 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr3_-_105512939 | 0.18 |
ENSRNOT00000011773
|
Actc1
|
actin, alpha, cardiac muscle 1 |
| chr3_+_171037957 | 0.18 |
ENSRNOT00000008764
|
Rbm38
|
RNA binding motif protein 38 |
| chr12_-_38340327 | 0.18 |
ENSRNOT00000001672
|
LOC100359550
|
protein S100-A11-like |
| chr11_+_73015387 | 0.17 |
ENSRNOT00000090978
|
Gp5
|
glycoprotein V (platelet) |
| chr9_+_10305470 | 0.17 |
ENSRNOT00000072592
|
Nrtn
|
neurturin |
| chr15_-_29369504 | 0.17 |
ENSRNOT00000060297
|
AABR07017624.1
|
|
| chr4_-_155401480 | 0.17 |
ENSRNOT00000020735
|
Apobec1
|
apolipoprotein B mRNA editing enzyme catalytic subunit 1 |
| chr12_+_24761210 | 0.17 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr1_+_81499821 | 0.17 |
ENSRNOT00000027100
|
Lypd3
|
Ly6/Plaur domain containing 3 |
| chr20_+_5008508 | 0.16 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr2_-_187863503 | 0.16 |
ENSRNOT00000093036
ENSRNOT00000026705 ENSRNOT00000082174 |
Lmna
|
lamin A/C |
| chr10_+_69423086 | 0.16 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr1_-_261446570 | 0.16 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr1_-_89068348 | 0.16 |
ENSRNOT00000033621
|
Upk1a
|
uroplakin 1A |
| chr1_+_199941161 | 0.16 |
ENSRNOT00000027616
|
Bag3
|
Bcl2-associated athanogene 3 |
| chr4_+_84423653 | 0.15 |
ENSRNOT00000012655
|
Chn2
|
chimerin 2 |
| chr16_-_19918644 | 0.15 |
ENSRNOT00000083345
ENSRNOT00000023926 |
Plvap
|
plasmalemma vesicle associated protein |
| chr10_-_88551056 | 0.15 |
ENSRNOT00000032737
|
Zfp385c
|
zinc finger protein 385C |
| chr12_+_38965063 | 0.15 |
ENSRNOT00000074936
|
Morn3
|
MORN repeat containing 3 |
| chr10_-_82229140 | 0.15 |
ENSRNOT00000004400
|
Epn3
|
epsin 3 |
| chr10_-_87232723 | 0.14 |
ENSRNOT00000015150
|
Krt25
|
keratin 25 |
| chr10_+_15088935 | 0.14 |
ENSRNOT00000030273
|
Gng13
|
G protein subunit gamma 13 |
| chr10_+_105499569 | 0.14 |
ENSRNOT00000088457
|
Sphk1
|
sphingosine kinase 1 |
| chr14_-_83776863 | 0.14 |
ENSRNOT00000026481
|
Smtn
|
smoothelin |
| chr18_+_29191731 | 0.14 |
ENSRNOT00000068085
ENSRNOT00000025495 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr12_+_48789261 | 0.13 |
ENSRNOT00000000893
|
Cmklr1
|
chemerin chemokine-like receptor 1 |
| chr3_+_147609095 | 0.13 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr1_-_88881460 | 0.13 |
ENSRNOT00000028287
|
Hcst
|
hematopoietic cell signal transducer |
| chr12_-_30314519 | 0.13 |
ENSRNOT00000001220
|
Zbed5
|
zinc finger, BED-type containing 5 |
| chr19_+_55669626 | 0.13 |
ENSRNOT00000033352
|
Cdh15
|
cadherin 15 |
| chr1_-_89560719 | 0.13 |
ENSRNOT00000028653
|
Scn1b
|
sodium voltage-gated channel beta subunit 1 |
| chr15_+_45834302 | 0.13 |
ENSRNOT00000051307
|
Serpine3
|
serpin family E member 3 |
| chr2_-_45518502 | 0.13 |
ENSRNOT00000014627
|
Hspb3
|
heat shock protein family B (small) member 3 |
| chr7_+_28654733 | 0.13 |
ENSRNOT00000006174
|
Pmch
|
pro-melanin-concentrating hormone |
| chr20_+_5040337 | 0.12 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr10_+_23914894 | 0.12 |
ENSRNOT00000071435
|
Ebf1
|
early B-cell factor 1 |
| chr8_-_45215974 | 0.12 |
ENSRNOT00000010792
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr1_+_267607416 | 0.12 |
ENSRNOT00000087894
ENSRNOT00000076367 ENSRNOT00000016851 |
Gsto2
Gsto1
|
glutathione S-transferase omega 2 glutathione S-transferase omega 1 |
| chr11_+_46179940 | 0.12 |
ENSRNOT00000088152
|
Tfg
|
Trk-fused gene |
| chr10_-_18063391 | 0.12 |
ENSRNOT00000072297
|
Fgf18
|
fibroblast growth factor 18 |
| chr19_-_645937 | 0.12 |
ENSRNOT00000016521
|
Car7
|
carbonic anhydrase 7 |
| chr5_-_155772040 | 0.12 |
ENSRNOT00000036788
|
Cela3b
|
chymotrypsin-like elastase family, member 3B |
| chr1_+_198690794 | 0.12 |
ENSRNOT00000023999
|
Zfp771
|
zinc finger protein 771 |
| chr1_-_213907144 | 0.12 |
ENSRNOT00000054874
|
Sigirr
|
single Ig and TIR domain containing |
| chr19_-_37938857 | 0.12 |
ENSRNOT00000026730
|
Slc12a4
|
solute carrier family 12 member 4 |
| chr10_-_13780283 | 0.12 |
ENSRNOT00000084200
|
AC103090.1
|
|
| chr1_-_13719829 | 0.12 |
ENSRNOT00000079522
|
Hebp2
|
heme binding protein 2 |
| chr1_-_101514547 | 0.12 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr18_-_3676188 | 0.11 |
ENSRNOT00000073811
|
Ankrd29
|
ankyrin repeat domain 29 |
| chr4_-_160662974 | 0.11 |
ENSRNOT00000089199
|
Tspan9
|
tetraspanin 9 |
| chrM_+_7758 | 0.11 |
ENSRNOT00000046201
|
Mt-atp8
|
mitochondrially encoded ATP synthase 8 |
| chr8_+_2635851 | 0.11 |
ENSRNOT00000061973
|
Casp4
|
caspase 4 |
| chr1_+_83003841 | 0.11 |
ENSRNOT00000057384
|
Ceacam4
|
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr1_+_31531143 | 0.11 |
ENSRNOT00000034704
|
Lrrc14b
|
leucine rich repeat containing 14B |
| chr4_-_10269748 | 0.11 |
ENSRNOT00000074662
|
Fam185a
|
family with sequence similarity 185, member A |
| chr1_+_82089922 | 0.11 |
ENSRNOT00000071251
|
Zfp526
|
zinc finger protein 526 |
| chr16_-_75648538 | 0.11 |
ENSRNOT00000048414
|
Defb14
|
defensin beta 14 |
| chr5_+_117052260 | 0.11 |
ENSRNOT00000010211
|
Patj
|
PATJ, crumbs cell polarity complex component |
| chr20_-_14020007 | 0.11 |
ENSRNOT00000093521
|
Ggt1
|
gamma-glutamyltransferase 1 |
| chr12_-_22006533 | 0.11 |
ENSRNOT00000059593
|
LOC100362783
|
Uncharacterized protein C7orf61 homolog |
| chr5_-_151459037 | 0.11 |
ENSRNOT00000064472
ENSRNOT00000087836 |
Sytl1
|
synaptotagmin-like 1 |
| chr1_+_99762253 | 0.10 |
ENSRNOT00000065876
|
Klk6
|
kallikrein related-peptidase 6 |
| chr7_-_3342491 | 0.10 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr20_+_6356423 | 0.10 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr19_-_37528011 | 0.10 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr11_-_33003021 | 0.10 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr10_+_103396155 | 0.10 |
ENSRNOT00000086924
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr7_-_123088819 | 0.10 |
ENSRNOT00000056077
|
AC096601.1
|
|
| chr3_+_1452644 | 0.10 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr4_+_169147243 | 0.10 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr5_+_8459660 | 0.10 |
ENSRNOT00000007491
|
Cpa6
|
carboxypeptidase A6 |
| chr20_-_3818045 | 0.10 |
ENSRNOT00000091622
|
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr1_-_89473904 | 0.10 |
ENSRNOT00000089474
|
Fxyd5
|
FXYD domain-containing ion transport regulator 5 |
| chr4_+_155313671 | 0.10 |
ENSRNOT00000020812
|
Mfap5
|
microfibrillar associated protein 5 |
| chr10_-_45534570 | 0.10 |
ENSRNOT00000058362
|
Gjc2
|
gap junction protein, gamma 2 |
| chr8_+_70915953 | 0.10 |
ENSRNOT00000046914
|
Rasl12
|
RAS-like, family 12 |
| chr9_-_101388833 | 0.10 |
ENSRNOT00000071817
|
Chchd2
|
coiled-coil-helix-coiled-coil-helix domain containing 2 |
| chr5_+_139597731 | 0.10 |
ENSRNOT00000072427
|
Cited4
|
Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 4 |
| chr2_-_53185926 | 0.10 |
ENSRNOT00000081558
|
Ghr
|
growth hormone receptor |
| chr10_-_45923829 | 0.10 |
ENSRNOT00000035285
|
Olr1462
|
olfactory receptor 1462 |
| chr17_-_70548071 | 0.10 |
ENSRNOT00000073144
|
Il2ra
|
interleukin 2 receptor subunit alpha |
| chr4_+_66276835 | 0.10 |
ENSRNOT00000007544
|
Fmc1
|
formation of mitochondrial complex V assembly factor 1 |
| chr13_-_90839411 | 0.10 |
ENSRNOT00000010594
|
Slamf9
|
SLAM family member 9 |
| chr5_+_129756149 | 0.09 |
ENSRNOT00000067277
|
Dmrta2
|
DMRT-like family A2 |
| chr17_-_78561613 | 0.09 |
ENSRNOT00000020220
|
Fam107b
|
family with sequence similarity 107, member B |
| chr13_-_50499060 | 0.09 |
ENSRNOT00000065347
ENSRNOT00000076924 |
Etnk2
|
ethanolamine kinase 2 |
| chr14_+_16491573 | 0.09 |
ENSRNOT00000002995
|
Sowahb
|
sosondowah ankyrin repeat domain family member B |
| chr1_+_238222521 | 0.09 |
ENSRNOT00000024000
|
Aldh1a1
|
aldehyde dehydrogenase 1 family, member A1 |
| chr8_+_117117430 | 0.09 |
ENSRNOT00000073247
|
Gpx1
|
glutathione peroxidase 1 |
| chr4_+_88271061 | 0.09 |
ENSRNOT00000087374
|
Vom1r86
|
vomeronasal 1 receptor 86 |
| chr5_-_135859643 | 0.09 |
ENSRNOT00000024719
|
Urod
|
uroporphyrinogen decarboxylase |
| chr12_+_24651314 | 0.09 |
ENSRNOT00000077016
ENSRNOT00000071569 |
Vps37d
|
VPS37D, ESCRT-I subunit |
| chr19_+_55381565 | 0.09 |
ENSRNOT00000018923
|
Cdt1
|
chromatin licensing and DNA replication factor 1 |
| chr1_+_242572533 | 0.09 |
ENSRNOT00000035123
|
Tmem252
|
transmembrane protein 252 |
| chr1_+_71434819 | 0.09 |
ENSRNOT00000059069
|
Zfp787
|
zinc finger protein 787 |
| chr5_+_153835821 | 0.09 |
ENSRNOT00000077610
|
Stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chr4_-_170912629 | 0.09 |
ENSRNOT00000055691
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr1_+_78820262 | 0.09 |
ENSRNOT00000022441
|
Gng8
|
G protein subunit gamma 8 |
| chr7_+_11769400 | 0.09 |
ENSRNOT00000044417
|
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr20_-_4921348 | 0.09 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chr1_-_87191588 | 0.09 |
ENSRNOT00000027998
|
LOC100361913
|
rCG54286-like |
| chr1_+_84573496 | 0.09 |
ENSRNOT00000057158
|
RGD1565183
|
similar to ribosomal protein L28 |
| chr10_+_56662242 | 0.09 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr9_-_16519144 | 0.09 |
ENSRNOT00000072581
|
LOC681367
|
hypothetical protein LOC681367 |
| chr12_+_25497104 | 0.09 |
ENSRNOT00000002028
|
Ncf1
|
neutrophil cytosolic factor 1 |
| chr2_+_193866951 | 0.09 |
ENSRNOT00000013393
|
S100a11
|
S100 calcium binding protein A11 |
| chr17_+_12261102 | 0.09 |
ENSRNOT00000015525
|
Nfil3
|
nuclear factor, interleukin 3 regulated |
| chr11_+_30904733 | 0.09 |
ENSRNOT00000036027
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr5_+_162031722 | 0.09 |
ENSRNOT00000020483
|
Lrrc38
|
leucine rich repeat containing 38 |
| chr2_-_195908037 | 0.08 |
ENSRNOT00000079776
|
Tuft1
|
tuftelin 1 |
| chr7_+_117951154 | 0.08 |
ENSRNOT00000060181
|
Znf7
|
zinc finger protein 7 |
| chr10_+_69412017 | 0.08 |
ENSRNOT00000009448
|
Ccl2
|
C-C motif chemokine ligand 2 |
| chr10_-_87067456 | 0.08 |
ENSRNOT00000014163
|
Ccr7
|
C-C motif chemokine receptor 7 |
| chr19_+_14835822 | 0.08 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr9_-_43116521 | 0.08 |
ENSRNOT00000039437
|
Ankrd23
|
ankyrin repeat domain 23 |
| chr20_-_4391402 | 0.08 |
ENSRNOT00000090518
ENSRNOT00000083489 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr3_+_5709236 | 0.08 |
ENSRNOT00000061201
ENSRNOT00000070887 |
Dbh
|
dopamine beta-hydroxylase |
| chr18_-_60002529 | 0.08 |
ENSRNOT00000081014
ENSRNOT00000024264 ENSRNOT00000059162 |
Nars
|
asparaginyl-tRNA synthetase |
| chr13_+_47602692 | 0.08 |
ENSRNOT00000038822
|
Fcmr
|
Fc fragment of IgM receptor |
| chr3_-_5575136 | 0.08 |
ENSRNOT00000008034
|
Slc2a6
|
solute carrier family 2 member 6 |
| chr7_-_92996025 | 0.08 |
ENSRNOT00000076327
|
Samd12
|
sterile alpha motif domain containing 12 |
| chr5_-_134526089 | 0.08 |
ENSRNOT00000013321
|
Cyp4b1
|
cytochrome P450, family 4, subfamily b, polypeptide 1 |
| chr2_-_205212681 | 0.08 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr10_+_10889488 | 0.08 |
ENSRNOT00000071746
|
Ubald1
|
UBA-like domain containing 1 |
| chr15_-_44860604 | 0.08 |
ENSRNOT00000018637
|
Nefm
|
neurofilament medium |
| chr10_+_103395511 | 0.08 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr1_-_216703534 | 0.08 |
ENSRNOT00000027972
|
Phlda2
|
pleckstrin homology-like domain, family A, member 2 |
| chr5_+_151776004 | 0.08 |
ENSRNOT00000009683
|
Nr0b2
|
nuclear receptor subfamily 0, group B, member 2 |
| chr20_+_4966817 | 0.08 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr12_+_16988136 | 0.08 |
ENSRNOT00000078925
ENSRNOT00000036744 |
Micall2
|
MICAL-like 2 |
| chr10_-_14703668 | 0.08 |
ENSRNOT00000024767
|
Tpsab1
|
tryptase alpha/beta 1 |
| chr1_+_91152635 | 0.08 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr20_+_4043741 | 0.08 |
ENSRNOT00000000525
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr1_+_84293102 | 0.08 |
ENSRNOT00000028468
|
Sertad1
|
SERTA domain containing 1 |
| chr8_+_132241134 | 0.08 |
ENSRNOT00000006055
|
Clec3b
|
C-type lectin domain family 3, member B |
| chr3_+_37545238 | 0.08 |
ENSRNOT00000070792
|
Tnfaip6
|
TNF alpha induced protein 6 |
| chr9_+_9961021 | 0.08 |
ENSRNOT00000075767
|
Tubb4a
|
tubulin, beta 4A class IVa |
| chr8_-_132027006 | 0.08 |
ENSRNOT00000050963
|
LOC367195
|
similar to 60S RIBOSOMAL PROTEIN L7 |
| chr4_+_98457810 | 0.08 |
ENSRNOT00000074175
|
AABR07060872.1
|
|
| chr13_-_109849632 | 0.08 |
ENSRNOT00000005085
|
Atf3
|
activating transcription factor 3 |
| chr16_+_20555395 | 0.08 |
ENSRNOT00000026652
|
Gdf15
|
growth differentiation factor 15 |
| chr19_-_37916813 | 0.08 |
ENSRNOT00000026585
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr1_-_24191908 | 0.08 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr8_+_116754178 | 0.08 |
ENSRNOT00000068295
ENSRNOT00000084429 |
Uba7
|
ubiquitin-like modifier activating enzyme 7 |
| chrX_-_14890606 | 0.08 |
ENSRNOT00000049864
|
RGD1560784
|
similar to RIKEN cDNA B630019K06 |
| chr1_+_175445088 | 0.08 |
ENSRNOT00000036718
|
Adm
|
adrenomedullin |
| chr2_-_149088787 | 0.08 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chr9_-_82154266 | 0.08 |
ENSRNOT00000024283
|
Cryba2
|
crystallin, beta A2 |
| chr1_+_89008117 | 0.08 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr12_-_2661565 | 0.08 |
ENSRNOT00000064859
|
Cd209f
|
CD209f antigen |
| chr1_+_78417719 | 0.08 |
ENSRNOT00000020609
|
Tmem160
|
transmembrane protein 160 |
| chr18_+_31094965 | 0.08 |
ENSRNOT00000026526
|
Rell2
|
RELT-like 2 |
| chr17_-_43675934 | 0.08 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr12_-_22478752 | 0.08 |
ENSRNOT00000089392
ENSRNOT00000086915 |
Ache
|
acetylcholinesterase |
| chr4_-_97784842 | 0.08 |
ENSRNOT00000007698
|
Gadd45a
|
growth arrest and DNA-damage-inducible, alpha |
| chr3_+_149624712 | 0.07 |
ENSRNOT00000018581
|
Bpifa1
|
BPI fold containing family A, member 1 |
| chr2_+_193572053 | 0.07 |
ENSRNOT00000056479
|
AABR07012329.1
|
|
| chr10_-_91122018 | 0.07 |
ENSRNOT00000080077
|
Dcakd
|
dephospho-CoA kinase domain containing |
| chr7_+_38819771 | 0.07 |
ENSRNOT00000006109
|
Lum
|
lumican |
| chr1_-_190596431 | 0.07 |
ENSRNOT00000079335
|
Pdzd9
|
PDZ domain containing 9 |
| chr10_-_89958805 | 0.07 |
ENSRNOT00000084266
|
Mpp3
|
membrane palmitoylated protein 3 |
| chr10_+_109786222 | 0.07 |
ENSRNOT00000054953
|
Npb
|
neuropeptide B |
| chr16_-_21089508 | 0.07 |
ENSRNOT00000072565
|
Hapln4
|
hyaluronan and proteoglycan link protein 4 |
| chr1_-_214455039 | 0.07 |
ENSRNOT00000071028
|
Polr2l
|
RNA polymerase II subunit L |
| chr1_-_206394346 | 0.07 |
ENSRNOT00000038122
|
LOC100302465
|
hypothetical LOC100302465 |
| chr15_+_36898195 | 0.07 |
ENSRNOT00000076266
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr12_+_23473270 | 0.07 |
ENSRNOT00000001935
|
Sh2b2
|
SH2B adaptor protein 2 |
| chr15_+_18540913 | 0.07 |
ENSRNOT00000010545
|
Pdhb
|
pyruvate dehydrogenase (lipoamide) beta |
| chr8_+_69971778 | 0.07 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr17_-_77304530 | 0.07 |
ENSRNOT00000024362
|
Phyh
|
phytanoyl-CoA 2-hydroxylase |
| chr13_+_96195836 | 0.07 |
ENSRNOT00000042547
|
RGD1563812
|
similar to basic transcription factor 3 |
| chr4_+_5841998 | 0.07 |
ENSRNOT00000010025
|
Xrcc2
|
X-ray repair cross complementing 2 |
| chr7_+_118490560 | 0.07 |
ENSRNOT00000064873
|
LOC108348145
|
zinc finger protein 7-like |
| chr1_-_222468896 | 0.07 |
ENSRNOT00000028754
|
Cox8a
|
cytochrome c oxidase subunit 8A |
| chr9_+_14951047 | 0.07 |
ENSRNOT00000074267
|
LOC100911489
|
uncharacterized LOC100911489 |
| chr5_+_153845094 | 0.07 |
ENSRNOT00000041600
|
Stpg1
|
sperm-tail PG-rich repeat containing 1 |
| chr7_-_2712723 | 0.07 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smarcc1_Fosl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0009080 | alanine catabolic process(GO:0006524) pyruvate family amino acid catabolic process(GO:0009080) D-amino acid metabolic process(GO:0046416) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.1 | 0.2 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.1 | 0.2 | GO:0060734 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.1 | 0.3 | GO:0009608 | response to symbiont(GO:0009608) |
| 0.0 | 0.1 | GO:0045079 | negative regulation of chemokine biosynthetic process(GO:0045079) |
| 0.0 | 0.2 | GO:0038128 | ERBB2 signaling pathway(GO:0038128) |
| 0.0 | 0.2 | GO:0030240 | skeletal muscle thin filament assembly(GO:0030240) |
| 0.0 | 0.1 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.0 | 0.2 | GO:0046087 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine to uridine editing(GO:0016554) cytidine metabolic process(GO:0046087) |
| 0.0 | 0.2 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.3 | GO:0035747 | natural killer cell chemotaxis(GO:0035747) regulation of natural killer cell chemotaxis(GO:2000501) |
| 0.0 | 0.1 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.2 | GO:0097070 | ductus arteriosus closure(GO:0097070) |
| 0.0 | 0.2 | GO:0032849 | positive regulation of cellular pH reduction(GO:0032849) |
| 0.0 | 0.2 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.0 | 0.6 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0071663 | granzyme B production(GO:0071613) regulation of granzyme B production(GO:0071661) positive regulation of granzyme B production(GO:0071663) |
| 0.0 | 0.1 | GO:2000417 | negative regulation of eosinophil migration(GO:2000417) |
| 0.0 | 0.1 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:1904849 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:0044107 | cellular alcohol metabolic process(GO:0044107) cellular alcohol biosynthetic process(GO:0044108) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:1901873 | regulation of post-translational protein modification(GO:1901873) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.2 | GO:0060574 | intestinal epithelial cell maturation(GO:0060574) |
| 0.0 | 0.1 | GO:2000520 | regulation of immunological synapse formation(GO:2000520) |
| 0.0 | 0.1 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.0 | GO:0097332 | response to antipsychotic drug(GO:0097332) |
| 0.0 | 0.1 | GO:0070947 | neutrophil mediated killing of fungus(GO:0070947) |
| 0.0 | 0.1 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.1 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.1 | GO:0002018 | renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0090272 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) negative regulation of fibroblast growth factor production(GO:0090272) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.1 | GO:0035766 | cell chemotaxis to fibroblast growth factor(GO:0035766) endothelial cell chemotaxis to fibroblast growth factor(GO:0035768) regulation of cell chemotaxis to fibroblast growth factor(GO:1904847) regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000544) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast cell proliferation(GO:0060720) cell proliferation involved in embryonic placenta development(GO:0060722) spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.1 | GO:0046490 | isopentenyl diphosphate biosynthetic process(GO:0009240) isopentenyl diphosphate metabolic process(GO:0046490) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.1 | GO:0016598 | protein arginylation(GO:0016598) |
| 0.0 | 0.1 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.0 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0003192 | mitral valve formation(GO:0003192) |
| 0.0 | 0.1 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:1903984 | positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.1 | GO:0060315 | negative regulation of ryanodine-sensitive calcium-release channel activity(GO:0060315) |
| 0.0 | 0.0 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.0 | 0.1 | GO:0032788 | saturated monocarboxylic acid metabolic process(GO:0032788) unsaturated monocarboxylic acid metabolic process(GO:0032789) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.0 | 0.1 | GO:1903121 | enzyme active site formation via L-cysteine sulfinic acid(GO:0018323) primary alcohol biosynthetic process(GO:0034309) cellular response to glyoxal(GO:0036471) TRAIL receptor biosynthetic process(GO:0045557) regulation of TRAIL receptor biosynthetic process(GO:0045560) glycolate biosynthetic process(GO:0046295) regulation of TRAIL-activated apoptotic signaling pathway(GO:1903121) negative regulation of TRAIL-activated apoptotic signaling pathway(GO:1903122) regulation of pyrroline-5-carboxylate reductase activity(GO:1903167) positive regulation of pyrroline-5-carboxylate reductase activity(GO:1903168) regulation of tyrosine 3-monooxygenase activity(GO:1903176) positive regulation of tyrosine 3-monooxygenase activity(GO:1903178) L-dopa metabolic process(GO:1903184) L-dopa biosynthetic process(GO:1903185) glyoxal metabolic process(GO:1903189) regulation of L-dopa biosynthetic process(GO:1903195) positive regulation of L-dopa biosynthetic process(GO:1903197) regulation of L-dopa decarboxylase activity(GO:1903198) positive regulation of L-dopa decarboxylase activity(GO:1903200) positive regulation of cellular amino acid biosynthetic process(GO:2000284) |
| 0.0 | 0.1 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.5 | GO:0095500 | acetylcholine receptor signaling pathway(GO:0095500) signal transduction involved in cellular response to ammonium ion(GO:1903831) response to acetylcholine(GO:1905144) cellular response to acetylcholine(GO:1905145) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.0 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:0052651 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.0 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.1 | GO:0099550 | trans-synaptic signalling, modulating synaptic transmission(GO:0099550) |
| 0.0 | 0.0 | GO:0071349 | interleukin-12-mediated signaling pathway(GO:0035722) cellular response to interleukin-12(GO:0071349) |
| 0.0 | 0.1 | GO:1904646 | cellular response to beta-amyloid(GO:1904646) |
| 0.0 | 0.0 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.0 | GO:0001998 | angiotensin mediated vasoconstriction involved in regulation of systemic arterial blood pressure(GO:0001998) |
| 0.0 | 0.1 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.0 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.0 | GO:1904398 | response to kainic acid(GO:1904373) positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) positive regulation of neuromuscular junction development(GO:1904398) |
| 0.0 | 0.0 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.0 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0035609 | C-terminal protein deglutamylation(GO:0035609) |
| 0.0 | 0.1 | GO:0032914 | positive regulation of transforming growth factor beta1 production(GO:0032914) |
| 0.0 | 0.0 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.0 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:0071681 | cellular response to indole-3-methanol(GO:0071681) |
| 0.0 | 0.1 | GO:0032429 | regulation of phospholipase A2 activity(GO:0032429) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:0061709 | reticulophagy(GO:0061709) |
| 0.0 | 0.0 | GO:0009822 | lipid hydroxylation(GO:0002933) alkaloid catabolic process(GO:0009822) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.0 | GO:1903847 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.2 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0009249 | protein lipoylation(GO:0009249) |
| 0.0 | 0.0 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0010940 | positive regulation of necrotic cell death(GO:0010940) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0036233 | glycine import(GO:0036233) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.0 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.0 | 0.1 | GO:0097084 | vascular smooth muscle cell development(GO:0097084) |
| 0.0 | 0.1 | GO:0015816 | glycine transport(GO:0015816) |
| 0.0 | 0.0 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.0 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0072602 | interleukin-4 secretion(GO:0072602) |
| 0.0 | 0.0 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.0 | 0.1 | GO:0045647 | negative regulation of erythrocyte differentiation(GO:0045647) |
| 0.0 | 0.0 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.0 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:0006478 | peptidyl-tyrosine sulfation(GO:0006478) |
| 0.0 | 0.0 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0051122 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.0 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.0 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0002098 | tRNA wobble uridine modification(GO:0002098) |
| 0.0 | 0.0 | GO:0052314 | isoquinoline alkaloid metabolic process(GO:0033076) phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.0 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.0 | GO:0046864 | isoprenoid transport(GO:0046864) terpenoid transport(GO:0046865) |
| 0.0 | 0.0 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.0 | GO:0098759 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 0.0 | GO:0060830 | ciliary receptor clustering involved in smoothened signaling pathway(GO:0060830) |
| 0.0 | 0.1 | GO:0036016 | response to interleukin-3(GO:0036015) cellular response to interleukin-3(GO:0036016) |
| 0.0 | 0.1 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0070557 | PCNA-p21 complex(GO:0070557) |
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.1 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.2 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0044299 | C-fiber(GO:0044299) |
| 0.0 | 0.1 | GO:0097169 | NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.0 | GO:0014801 | longitudinal sarcoplasmic reticulum(GO:0014801) |
| 0.0 | 0.1 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0045261 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) proton-transporting ATP synthase complex, catalytic core F(1)(GO:0045261) |
| 0.0 | 0.0 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.0 | GO:0044214 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.0 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.0 | 0.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.1 | GO:0001652 | granular component(GO:0001652) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0042719 | mitochondrial intermembrane space protein transporter complex(GO:0042719) |
| 0.0 | 0.0 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.1 | 0.6 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.1 | 0.1 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 0.1 | 0.2 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.1 | 0.2 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.1 | 0.2 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.0 | 0.6 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.6 | GO:0030548 | acetylcholine receptor regulator activity(GO:0030548) neurotransmitter receptor regulator activity(GO:0099602) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.2 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.1 | GO:0030614 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.6 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.1 | GO:0004903 | growth hormone receptor activity(GO:0004903) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.0 | GO:0035373 | chondroitin sulfate proteoglycan binding(GO:0035373) |
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0098603 | selenol Se-methyltransferase activity(GO:0098603) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.1 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.1 | GO:0036470 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.0 | 0.1 | GO:0001733 | galactosylceramide sulfotransferase activity(GO:0001733) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.3 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.0 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.0 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.0 | 0.2 | GO:0004862 | cAMP-dependent protein kinase inhibitor activity(GO:0004862) |
| 0.0 | 0.0 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.0 | 0.4 | GO:0016676 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.2 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.0 | GO:0004775 | succinate-CoA ligase activity(GO:0004774) succinate-CoA ligase (ADP-forming) activity(GO:0004775) |
| 0.0 | 0.1 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.0 | GO:0004058 | aromatic-L-amino-acid decarboxylase activity(GO:0004058) L-dopa decarboxylase activity(GO:0036468) |
| 0.0 | 0.1 | GO:0036374 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.1 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.3 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.1 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.0 | GO:0035514 | DNA demethylase activity(GO:0035514) oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0003865 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.0 | 0.1 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.0 | GO:0032551 | UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.0 | GO:0005143 | interleukin-12 receptor binding(GO:0005143) |
| 0.0 | 0.0 | GO:0008476 | protein-tyrosine sulfotransferase activity(GO:0008476) |
| 0.0 | 0.2 | GO:0004602 | glutathione peroxidase activity(GO:0004602) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.2 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.0 | GO:0004457 | lactate dehydrogenase activity(GO:0004457) L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.0 | GO:0019797 | procollagen-proline 3-dioxygenase activity(GO:0019797) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.0 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.0 | GO:0008254 | 3'-nucleotidase activity(GO:0008254) |
| 0.0 | 0.4 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.1 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.1 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.1 | GO:0015187 | glycine transmembrane transporter activity(GO:0015187) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.0 | PID SYNDECAN 1 PATHWAY | Syndecan-1-mediated signaling events |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
| 0.0 | 0.0 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.0 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.0 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.0 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.1 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.4 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.3 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |