Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
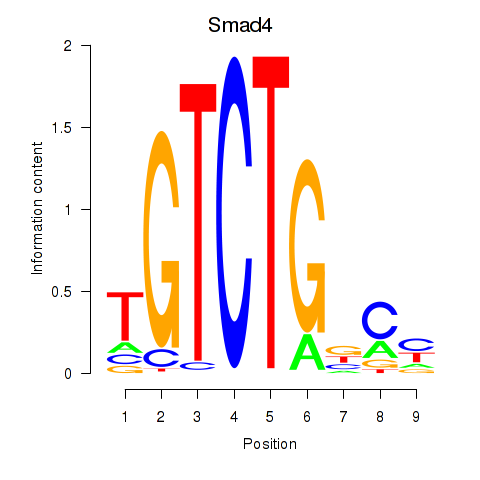
Results for Smad4
Z-value: 0.61
Transcription factors associated with Smad4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Smad4
|
ENSRNOG00000051965 | SMAD family member 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smad4 | rn6_v1_chr18_-_69657301_69657301 | 0.26 | 6.7e-01 | Click! |
Activity profile of Smad4 motif
Sorted Z-values of Smad4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_88670430 | 1.25 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr8_-_98738446 | 0.41 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr17_-_9695292 | 0.41 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr4_+_84194347 | 0.36 |
ENSRNOT00000084256
|
Chn2
|
chimerin 2 |
| chr12_-_41671437 | 0.34 |
ENSRNOT00000001883
|
Lhx5
|
LIM homeobox 5 |
| chr2_-_198360678 | 0.27 |
ENSRNOT00000051917
|
Hist2h2ac
|
histone cluster 2 H2A family member c |
| chr5_-_152473868 | 0.25 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr7_-_70842405 | 0.20 |
ENSRNOT00000047449
|
Nxph4
|
neurexophilin 4 |
| chr7_+_123168811 | 0.18 |
ENSRNOT00000007091
|
Csdc2
|
cold shock domain containing C2 |
| chr17_-_43821536 | 0.17 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr1_-_101856881 | 0.17 |
ENSRNOT00000028615
|
Grin2d
|
glutamate ionotropic receptor NMDA type subunit 2D |
| chr10_-_45297385 | 0.16 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr4_+_153774486 | 0.16 |
ENSRNOT00000074096
|
Tuba8
|
tubulin, alpha 8 |
| chr5_-_152464850 | 0.15 |
ENSRNOT00000021937
|
Zfp593
|
zinc finger protein 593 |
| chr17_-_44744902 | 0.14 |
ENSRNOT00000085381
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr3_+_56862691 | 0.10 |
ENSRNOT00000087712
|
Gad1
|
glutamate decarboxylase 1 |
| chr8_+_58347736 | 0.09 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr14_-_35149608 | 0.08 |
ENSRNOT00000003050
ENSRNOT00000090654 |
Kit
|
KIT proto-oncogene receptor tyrosine kinase |
| chr13_-_91872954 | 0.08 |
ENSRNOT00000004613
ENSRNOT00000079263 |
Cadm3
|
cell adhesion molecule 3 |
| chr17_+_78739389 | 0.08 |
ENSRNOT00000020384
|
LOC102547897
|
uncharacterized LOC102547897 |
| chr12_+_37668369 | 0.08 |
ENSRNOT00000001419
|
Cdk2ap1
|
cyclin-dependent kinase 2 associated protein 1 |
| chr14_-_34218961 | 0.07 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chr4_-_170145262 | 0.07 |
ENSRNOT00000074526
|
Hist1h4b
|
histone cluster 1, H4b |
| chr10_-_107539658 | 0.07 |
ENSRNOT00000089346
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr1_+_72636959 | 0.07 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chr1_+_219345918 | 0.07 |
ENSRNOT00000025018
|
Cdk2ap2
|
cyclin-dependent kinase 2 associated protein 2 |
| chr13_+_106463368 | 0.06 |
ENSRNOT00000003489
|
Esrrg
|
estrogen-related receptor gamma |
| chr1_+_14224393 | 0.06 |
ENSRNOT00000016037
|
Perp
|
PERP, TP53 apoptosis effector |
| chr17_-_43689311 | 0.06 |
ENSRNOT00000028779
|
Hist1h2bcl1
|
histone cluster 1, H2bc-like 1 |
| chr4_+_51553454 | 0.06 |
ENSRNOT00000073466
|
LOC100909784
|
leiomodin-2-like |
| chr13_+_48287873 | 0.06 |
ENSRNOT00000068223
|
Fam72a
|
family with sequence similarity 72, member A |
| chr9_+_94562839 | 0.05 |
ENSRNOT00000022308
|
RGD1311447
|
LOC363276 |
| chr10_-_107539465 | 0.05 |
ENSRNOT00000004524
|
Rbfox3
|
RNA binding protein, fox-1 homolog 3 |
| chr1_-_101514547 | 0.05 |
ENSRNOT00000079633
|
Ppp1r15a
|
protein phosphatase 1, regulatory subunit 15A |
| chr16_+_61758917 | 0.05 |
ENSRNOT00000084858
|
Leprotl1
|
leptin receptor overlapping transcript-like 1 |
| chr10_+_56453877 | 0.05 |
ENSRNOT00000031640
|
Plscr3
|
phospholipid scramblase 3 |
| chr17_-_44520240 | 0.05 |
ENSRNOT00000086538
|
Hist1h2bh
|
histone cluster 1 H2B family member h |
| chr15_+_48327461 | 0.05 |
ENSRNOT00000018071
|
Ints9
|
integrator complex subunit 9 |
| chr17_-_43770561 | 0.04 |
ENSRNOT00000088408
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr12_+_21767606 | 0.04 |
ENSRNOT00000059602
|
LOC103691013
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr3_+_148541909 | 0.04 |
ENSRNOT00000012187
|
Ccm2l
|
CCM2 like scaffolding protein |
| chr9_-_16647458 | 0.04 |
ENSRNOT00000024380
|
Mrpl2
|
mitochondrial ribosomal protein L2 |
| chrX_+_106823491 | 0.04 |
ENSRNOT00000045997
|
Bex3
|
brain expressed X-linked 3 |
| chr7_-_14418950 | 0.03 |
ENSRNOT00000066722
|
Rasal3
|
RAS protein activator like 3 |
| chr7_+_119647375 | 0.03 |
ENSRNOT00000046563
|
Kctd17
|
potassium channel tetramerization domain containing 17 |
| chr8_+_117280705 | 0.03 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr10_-_90265017 | 0.03 |
ENSRNOT00000064283
ENSRNOT00000048418 |
Ubtf
|
upstream binding transcription factor, RNA polymerase I |
| chr17_+_44520537 | 0.03 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chr15_-_88622413 | 0.03 |
ENSRNOT00000092174
|
Pou4f1
|
POU class 4 homeobox 1 |
| chr9_+_66335492 | 0.03 |
ENSRNOT00000037555
|
RGD1562029
|
similar to KIAA2012 protein |
| chr17_+_23135985 | 0.03 |
ENSRNOT00000090794
|
Nedd9
|
neural precursor cell expressed, developmentally down-regulated 9 |
| chr14_+_91557601 | 0.03 |
ENSRNOT00000038733
|
RGD1309870
|
hypothetical LOC289778 |
| chr9_+_117583610 | 0.03 |
ENSRNOT00000088647
ENSRNOT00000049426 |
Epb41l3
|
erythrocyte membrane protein band 4.1-like 3 |
| chr6_+_108745895 | 0.02 |
ENSRNOT00000061786
|
LOC108348062
|
eukaryotic translation initiation factor 3 subunit H |
| chr9_+_16647598 | 0.02 |
ENSRNOT00000087413
|
Klc4
|
kinesin light chain 4 |
| chr9_+_16647922 | 0.02 |
ENSRNOT00000031625
|
Klc4
|
kinesin light chain 4 |
| chr12_+_4546287 | 0.02 |
ENSRNOT00000001416
|
Higd2al1
|
HIG1 hypoxia inducible domain family, member 2A-like 1 |
| chr2_+_209839299 | 0.02 |
ENSRNOT00000092450
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr7_-_143217535 | 0.02 |
ENSRNOT00000088316
|
Kb15
|
type II keratin Kb15 |
| chr20_-_2211995 | 0.01 |
ENSRNOT00000077201
|
Trim26
|
tripartite motif-containing 26 |
| chr15_+_23792931 | 0.01 |
ENSRNOT00000092091
|
Samd4a
|
sterile alpha motif domain containing 4A |
| chr10_-_34333305 | 0.01 |
ENSRNOT00000071365
|
Olr1384
|
olfactory receptor gene Olr1384 |
| chrX_+_107370431 | 0.01 |
ENSRNOT00000044372
|
Tceal1
|
transcription elongation factor A like 1 |
| chrX_+_21696772 | 0.01 |
ENSRNOT00000043559
|
Hsd17b10
|
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr2_+_209838869 | 0.01 |
ENSRNOT00000092365
|
Kcna2
|
potassium voltage-gated channel subfamily A member 2 |
| chr3_-_113342675 | 0.01 |
ENSRNOT00000020130
|
Strc
|
stereocilin |
| chr3_-_8979889 | 0.00 |
ENSRNOT00000065128
|
Crat
|
carnitine O-acetyltransferase |
| chr10_+_46940965 | 0.00 |
ENSRNOT00000005366
|
Llgl1
|
LLGL1, scribble cell polarity complex component |
| chr7_-_120403523 | 0.00 |
ENSRNOT00000015283
|
Sox10
|
SRY box 10 |
| chr7_+_144623555 | 0.00 |
ENSRNOT00000022217
|
Hoxc6
|
homeo box C6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Smad4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.2 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.3 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.4 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.4 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.1 | GO:1905065 | Kit signaling pathway(GO:0038109) melanocyte migration(GO:0097324) positive regulation of vascular smooth muscle cell differentiation(GO:1905065) |
| 0.0 | 0.1 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.0 | 0.1 | GO:1902310 | regulation of endoplasmic reticulum stress-induced eIF2 alpha phosphorylation(GO:0060734) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) response to environmental enrichment(GO:0090648) positive regulation of peptidyl-serine dephosphorylation(GO:1902310) |
| 0.0 | 0.0 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.0 | GO:0051140 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | GO:0000788 | nuclear nucleosome(GO:0000788) |
| 0.0 | 1.2 | GO:0005791 | rough endoplasmic reticulum(GO:0005791) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0001565 | phorbol ester receptor activity(GO:0001565) |
| 0.0 | 0.1 | GO:0031177 | phosphopantetheine binding(GO:0031177) |
| 0.0 | 1.2 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.2 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME RNA POL I PROMOTER OPENING | Genes involved in RNA Polymerase I Promoter Opening |