Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Six6
Z-value: 0.59
Transcription factors associated with Six6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Six6
|
ENSRNOG00000006296 | SIX homeobox 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six6 | rn6_v1_chr6_+_95816749_95816749 | 0.05 | 9.4e-01 | Click! |
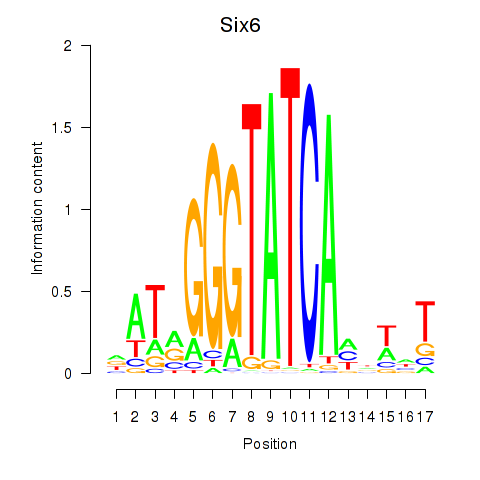
Activity profile of Six6 motif
Sorted Z-values of Six6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_57516347 | 0.34 |
ENSRNOT00000082215
|
Gm9949
|
predicted gene 9949 |
| chr4_-_80395502 | 0.30 |
ENSRNOT00000014437
|
Npvf
|
neuropeptide VF precursor |
| chr1_-_213650247 | 0.25 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chr10_+_31647898 | 0.18 |
ENSRNOT00000066536
|
LOC689968
|
similar to kidney injury molecule 1 |
| chr8_+_69753373 | 0.17 |
ENSRNOT00000066973
|
Tipin
|
timeless interacting protein |
| chr1_-_214252456 | 0.17 |
ENSRNOT00000023504
|
Irf7
|
interferon regulatory factor 7 |
| chr3_-_40477754 | 0.16 |
ENSRNOT00000091168
|
AABR07052166.1
|
|
| chr5_+_122508388 | 0.16 |
ENSRNOT00000038410
|
Tctex1d1
|
Tctex1 domain containing 1 |
| chr1_-_198104109 | 0.16 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr9_+_53440272 | 0.15 |
ENSRNOT00000059159
|
LOC685203
|
hypothetical protein LOC685203 |
| chr16_-_81243221 | 0.13 |
ENSRNOT00000076763
|
Gas6
|
growth arrest specific 6 |
| chr18_-_40716686 | 0.13 |
ENSRNOT00000000172
|
Cdo1
|
cysteine dioxygenase type 1 |
| chr1_-_225830300 | 0.12 |
ENSRNOT00000072584
ENSRNOT00000027502 |
Scgb2a2
|
secretoglobin, family 2A, member 2 |
| chrX_-_37705263 | 0.12 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr3_-_59166356 | 0.12 |
ENSRNOT00000047713
|
AABR07052519.1
|
|
| chr5_-_134927235 | 0.12 |
ENSRNOT00000016751
|
Uqcrh
|
ubiquinol-cytochrome c reductase hinge protein |
| chr8_+_76426335 | 0.11 |
ENSRNOT00000085496
|
Gtf2a2
|
general transcription factor2A subunit 2 |
| chr7_+_140248852 | 0.11 |
ENSRNOT00000087338
|
RGD1305928
|
hypothetical LOC300207 |
| chr3_-_164728338 | 0.11 |
ENSRNOT00000035463
|
Fam65c
|
family with sequence similarity 65, member C |
| chr8_+_91368111 | 0.11 |
ENSRNOT00000083151
|
Ttk
|
Ttk protein kinase |
| chr4_-_156629949 | 0.11 |
ENSRNOT00000042249
|
Vom2r52
|
vomeronasal 2 receptor, 52 |
| chr5_+_146294030 | 0.11 |
ENSRNOT00000073637
|
AABR07049960.1
|
|
| chr8_-_96547568 | 0.11 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr10_+_42933077 | 0.11 |
ENSRNOT00000003361
|
Mfap3
|
microfibrillar-associated protein 3 |
| chr9_-_19978013 | 0.10 |
ENSRNOT00000038222
|
Pla2g7
|
phospholipase A2 group VII |
| chr10_+_88227360 | 0.10 |
ENSRNOT00000041033
|
Eif1
|
eukaryotic translation initiation factor 1 |
| chr16_+_70905993 | 0.10 |
ENSRNOT00000078964
|
AABR07026294.1
|
|
| chr12_-_20276121 | 0.10 |
ENSRNOT00000065873
|
LOC685157
|
similar to paired immunoglobin-like type 2 receptor beta |
| chr1_-_100530183 | 0.10 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr4_-_161091010 | 0.10 |
ENSRNOT00000043708
|
Senp17
|
Sumo1/sentrin/SMT3 specific peptidase 17 |
| chr19_+_15033108 | 0.10 |
ENSRNOT00000021812
|
Ces1d
|
carboxylesterase 1D |
| chr8_-_62386264 | 0.09 |
ENSRNOT00000026165
|
Cplx3
|
complexin 3 |
| chr4_-_164453171 | 0.09 |
ENSRNOT00000077539
ENSRNOT00000083610 ENSRNOT00000079975 |
Ly49s6
|
Ly49 stimulatory receptor 6 |
| chrX_+_74273182 | 0.09 |
ENSRNOT00000003922
|
Tsx
|
testis specific X-linked gene |
| chr16_+_49462889 | 0.09 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
| chr4_-_117531480 | 0.09 |
ENSRNOT00000021120
|
Nat8f5
|
N-acetyltransferase 8 (GCN5-related) family member 5 |
| chr4_+_88832178 | 0.09 |
ENSRNOT00000088983
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr3_+_103192683 | 0.09 |
ENSRNOT00000037551
|
LOC100363452
|
ribosomal protein S8-like |
| chrX_+_28435507 | 0.09 |
ENSRNOT00000005615
|
Prps2
|
phosphoribosyl pyrophosphate synthetase 2 |
| chr17_+_5311274 | 0.09 |
ENSRNOT00000067020
|
LOC102547665
|
spermatogenesis-associated protein 31D1-like |
| chr18_+_25613831 | 0.08 |
ENSRNOT00000091040
|
Tslp
|
thymic stromal lymphopoietin |
| chr7_-_117773134 | 0.08 |
ENSRNOT00000045135
|
Recql4
|
RecQ like helicase 4 |
| chr1_-_222167447 | 0.08 |
ENSRNOT00000028687
|
Prdx5
|
peroxiredoxin 5 |
| chr3_-_2309179 | 0.08 |
ENSRNOT00000012375
|
Noxa1
|
NADPH oxidase activator 1 |
| chr1_+_245237736 | 0.08 |
ENSRNOT00000035814
|
Vldlr
|
very low density lipoprotein receptor |
| chr9_+_95274707 | 0.08 |
ENSRNOT00000045163
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr2_-_196304169 | 0.08 |
ENSRNOT00000028639
|
Scnm1
|
sodium channel modifier 1 |
| chr12_-_21362205 | 0.08 |
ENSRNOT00000064787
|
LOC108348155
|
paired immunoglobulin-like type 2 receptor beta-2 |
| chr5_+_124442293 | 0.08 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr19_+_15195565 | 0.08 |
ENSRNOT00000090865
ENSRNOT00000078874 |
Ces1d
|
carboxylesterase 1D |
| chr15_+_32386816 | 0.07 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr11_+_44192940 | 0.07 |
ENSRNOT00000002255
|
St3gal6
|
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr1_+_215610368 | 0.07 |
ENSRNOT00000078903
ENSRNOT00000087781 |
Tnni2
|
troponin I2, fast skeletal type |
| chr1_+_88686731 | 0.07 |
ENSRNOT00000028222
|
Polr2i
|
RNA polymerase II subunit I |
| chr2_+_208750356 | 0.07 |
ENSRNOT00000041562
|
Chia
|
chitinase, acidic |
| chr1_+_147713892 | 0.07 |
ENSRNOT00000092985
ENSRNOT00000054742 ENSRNOT00000074103 |
Cyp2c6v1
|
cytochrome P450, family 2, subfamily C, polypeptide 6, variant 1 |
| chr15_+_41937880 | 0.07 |
ENSRNOT00000032514
|
Kcnrg
|
potassium channel regulator |
| chr1_-_88686615 | 0.07 |
ENSRNOT00000070921
|
Tbcb
|
tubulin folding cofactor B |
| chr2_+_92573004 | 0.07 |
ENSRNOT00000065774
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr8_+_33514042 | 0.07 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chrX_+_63542191 | 0.07 |
ENSRNOT00000073955
|
Apoo
|
apolipoprotein O |
| chr8_-_71052529 | 0.07 |
ENSRNOT00000033512
|
Ankdd1a
|
ankyrin repeat and death domain containing 1A |
| chr1_+_190914946 | 0.07 |
ENSRNOT00000039447
|
LOC691551
|
similar to F28B3.5a |
| chr15_+_17834635 | 0.07 |
ENSRNOT00000085530
|
LOC361016
|
similar to RIKEN cDNA 4933406L09 |
| chr2_+_27107318 | 0.07 |
ENSRNOT00000019553
|
Arhgef26
|
Rho guanine nucleotide exchange factor 26 |
| chr10_+_90984227 | 0.07 |
ENSRNOT00000003890
ENSRNOT00000093707 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr5_-_118541928 | 0.07 |
ENSRNOT00000012947
|
Itgb3bp
|
integrin subunit beta 3 binding protein |
| chr2_-_88660449 | 0.07 |
ENSRNOT00000051741
|
Slc7a12
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr3_-_9262628 | 0.07 |
ENSRNOT00000013286
|
Aif1l
|
allograft inflammatory factor 1-like |
| chr2_-_236431596 | 0.07 |
ENSRNOT00000055570
ENSRNOT00000089674 |
Cyp2u1
|
cytochrome P450, family 2, subfamily u, polypeptide 1 |
| chr1_+_99616447 | 0.07 |
ENSRNOT00000029197
|
Klk14
|
kallikrein related-peptidase 14 |
| chr7_-_120874308 | 0.06 |
ENSRNOT00000078320
|
Fam227a
|
family with sequence similarity 227, member A |
| chrX_+_112358748 | 0.06 |
ENSRNOT00000074776
|
Atg4a
|
autophagy related 4A, cysteine peptidase |
| chr4_-_9060554 | 0.06 |
ENSRNOT00000046248
|
AABR07059215.1
|
|
| chr2_+_5437067 | 0.06 |
ENSRNOT00000093516
|
Pou5f2
|
POU domain class 5, transcription factor 2 |
| chr20_-_28873363 | 0.06 |
ENSRNOT00000071064
|
Snrpd2l
|
small nuclear ribonucleoprotein D2-like |
| chr1_-_151106802 | 0.06 |
ENSRNOT00000021971
|
Tyr
|
tyrosinase |
| chr3_-_173944396 | 0.06 |
ENSRNOT00000079019
|
Sycp2
|
synaptonemal complex protein 2 |
| chr11_+_86421106 | 0.06 |
ENSRNOT00000002599
|
LOC100362453
|
serine/threonine-protein phosphatase 2A catalytic subunit alpha-like |
| chr2_+_208749996 | 0.06 |
ENSRNOT00000086321
|
Chia
|
chitinase, acidic |
| chr8_+_56585396 | 0.06 |
ENSRNOT00000085316
ENSRNOT00000034924 |
Rdx
|
radixin |
| chr7_+_29435444 | 0.06 |
ENSRNOT00000008613
|
Slc5a8
|
solute carrier family 5 member 8 |
| chr3_-_124879216 | 0.06 |
ENSRNOT00000028886
|
Tmem230
|
transmembrane protein 230 |
| chr1_+_66959610 | 0.06 |
ENSRNOT00000072122
ENSRNOT00000045994 |
Vom1r48
|
vomeronasal 1 receptor 48 |
| chr13_-_98023829 | 0.06 |
ENSRNOT00000075426
|
Kif28p
|
kinesin family member 28, pseudogene |
| chr18_-_27452420 | 0.06 |
ENSRNOT00000037888
|
Cdc23
|
cell division cycle 23 |
| chr7_+_114997103 | 0.06 |
ENSRNOT00000010224
|
Ptp4a3
|
protein tyrosine phosphatase type IVA, member 3 |
| chr8_+_32604365 | 0.06 |
ENSRNOT00000071210
|
AABR07069587.1
|
|
| chrX_+_19854836 | 0.06 |
ENSRNOT00000075010
|
AABR07037395.2
|
|
| chr4_-_181348799 | 0.06 |
ENSRNOT00000033743
|
LOC690784
|
hypothetical protein LOC690783 |
| chr5_+_118541979 | 0.06 |
ENSRNOT00000013089
|
Efcab7
|
EF-hand calcium binding domain 7 |
| chr2_+_211159062 | 0.06 |
ENSRNOT00000027099
|
Mybphl
|
myosin binding protein H-like |
| chr5_-_74190991 | 0.06 |
ENSRNOT00000090366
|
Epb41l4b
|
erythrocyte membrane protein band 4.1 like 4B |
| chrX_+_125314613 | 0.06 |
ENSRNOT00000029193
|
LOC691215
|
hypothetical protein LOC691215 |
| chr9_+_93086012 | 0.06 |
ENSRNOT00000089470
|
Psmd1
|
proteasome 26S subunit, non-ATPase 1 |
| chr1_-_229601032 | 0.06 |
ENSRNOT00000016690
|
Cntf
|
ciliary neurotrophic factor |
| chr1_-_167893061 | 0.06 |
ENSRNOT00000049401
|
Olr60
|
olfactory receptor 60 |
| chr8_-_46694613 | 0.05 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr13_-_103080920 | 0.05 |
ENSRNOT00000034990
|
AABR07022022.1
|
|
| chrX_-_142248369 | 0.05 |
ENSRNOT00000091330
|
Fgf13
|
fibroblast growth factor 13 |
| chr1_-_62114672 | 0.05 |
ENSRNOT00000072607
|
AABR07001923.1
|
|
| chr17_-_67945037 | 0.05 |
ENSRNOT00000023033
|
Klf6
|
Kruppel-like factor 6 |
| chr3_-_102826379 | 0.05 |
ENSRNOT00000073423
|
Olr769
|
olfactory receptor 769 |
| chr8_-_39190457 | 0.05 |
ENSRNOT00000090161
|
Stt3a
|
STT3A, catalytic subunit of the oligosaccharyltransferase complex |
| chr6_-_1942972 | 0.05 |
ENSRNOT00000048711
|
Cdc42ep3
|
CDC42 effector protein 3 |
| chr10_-_103972668 | 0.05 |
ENSRNOT00000004836
|
Atp5h
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit d |
| chr8_+_84945444 | 0.05 |
ENSRNOT00000008244
|
Klhl31
|
kelch-like family member 31 |
| chr9_+_23420654 | 0.05 |
ENSRNOT00000073595
|
LOC688459
|
hypothetical protein LOC688459 |
| chr10_+_104952237 | 0.05 |
ENSRNOT00000085222
|
RGD1559482
|
similar to immunoglobulin superfamily, member 7 |
| chr5_+_118743632 | 0.05 |
ENSRNOT00000013785
|
Pgm1
|
phosphoglucomutase 1 |
| chr8_-_49502647 | 0.05 |
ENSRNOT00000040313
|
Tmprss4
|
transmembrane protease, serine 4 |
| chr18_+_30010918 | 0.05 |
ENSRNOT00000084132
|
Pcdha4
|
protocadherin alpha 4 |
| chr2_+_189106039 | 0.05 |
ENSRNOT00000028210
|
Ube2q1
|
ubiquitin conjugating enzyme E2 Q1 |
| chr1_+_64480929 | 0.05 |
ENSRNOT00000087016
|
Olr1l
|
olfactory receptor 1-like |
| chr20_+_2094931 | 0.05 |
ENSRNOT00000001013
|
Ppp1r11
|
protein phosphatase 1, regulatory (inhibitor) subunit 11 |
| chr13_-_47916185 | 0.05 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr10_-_88144468 | 0.05 |
ENSRNOT00000084619
|
Ka11
|
type I keratin KA11 |
| chr10_-_85725429 | 0.05 |
ENSRNOT00000005471
|
Rpl23
|
ribosomal protein L23 |
| chr14_-_6533524 | 0.05 |
ENSRNOT00000079795
|
Abcg3l1
|
ATP-binding cassette, subfamily G (WHITE), member 3-like 1 |
| chr2_+_182006242 | 0.05 |
ENSRNOT00000064091
|
Fga
|
fibrinogen alpha chain |
| chr6_-_105160470 | 0.05 |
ENSRNOT00000008367
|
Adam4
|
a disintegrin and metalloprotease domain 4 |
| chr16_-_8350067 | 0.05 |
ENSRNOT00000026932
|
LOC100362432
|
mitochondrial import inner membrane translocase subunit Tim23-like |
| chr4_-_160803558 | 0.05 |
ENSRNOT00000049542
|
Senp18
|
Sumo1/sentrin/SMT3 specific peptidase 18 |
| chr5_-_169301728 | 0.05 |
ENSRNOT00000013646
|
Espn
|
espin |
| chr9_+_41045363 | 0.05 |
ENSRNOT00000066284
|
Cfc1
|
cripto, FRL-1, cryptic family 1 |
| chr13_-_95943761 | 0.05 |
ENSRNOT00000005961
|
Adss
|
adenylosuccinate synthase |
| chrX_-_128268285 | 0.05 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr1_-_281376834 | 0.05 |
ENSRNOT00000013031
|
Fam204a
|
family with sequence similarity 204, member A |
| chr1_-_167884690 | 0.05 |
ENSRNOT00000091372
|
Olr61
|
olfactory receptor 61 |
| chr1_-_190914610 | 0.05 |
ENSRNOT00000023189
|
Cdr2
|
cerebellar degeneration-related protein 2 |
| chr15_-_36114752 | 0.05 |
ENSRNOT00000045209
|
Gzmbl3
|
Granzyme B-like 3 |
| chr7_-_130350570 | 0.05 |
ENSRNOT00000055805
|
Odf3b
|
outer dense fiber of sperm tails 3B |
| chr8_-_6235967 | 0.05 |
ENSRNOT00000068290
|
LOC654482
|
hypothetical protein LOC654482 |
| chr4_+_152630469 | 0.05 |
ENSRNOT00000013721
|
Ninj2
|
ninjurin 2 |
| chr10_+_104952458 | 0.05 |
ENSRNOT00000074082
|
RGD1559482
|
similar to immunoglobulin superfamily, member 7 |
| chr10_+_103972888 | 0.05 |
ENSRNOT00000067838
|
Kctd2
|
potassium channel tetramerization domain containing 2 |
| chr7_-_119712888 | 0.05 |
ENSRNOT00000077438
|
Il2rb
|
interleukin 2 receptor subunit beta |
| chr12_+_19890749 | 0.05 |
ENSRNOT00000074970
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr9_-_113246545 | 0.05 |
ENSRNOT00000034759
|
Tmem232
|
transmembrane protein 232 |
| chr19_+_51317425 | 0.05 |
ENSRNOT00000019298
|
Cdh13
|
cadherin 13 |
| chr12_+_4248808 | 0.05 |
ENSRNOT00000042410
|
AABR07035089.1
|
|
| chr7_+_117643206 | 0.05 |
ENSRNOT00000077588
|
Adck5
|
aarF domain containing kinase 5 |
| chr3_+_119173818 | 0.05 |
ENSRNOT00000015124
|
Usp8
|
ubiquitin specific peptidase 8 |
| chr19_+_49792273 | 0.05 |
ENSRNOT00000017941
ENSRNOT00000083991 |
Cmip
|
c-Maf-inducing protein |
| chr1_-_60828260 | 0.04 |
ENSRNOT00000059671
|
Vom1r-ps112
|
vomeronasal 1 receptor pseudogene 112 |
| chr4_-_99810270 | 0.04 |
ENSRNOT00000078123
|
Ptcd3
|
Pentatricopeptide repeat domain 3 |
| chr1_+_229267916 | 0.04 |
ENSRNOT00000073717
ENSRNOT00000082670 ENSRNOT00000076941 |
LOC100910851
|
serine protease inhibitor Kazal-type 5-like |
| chr19_+_38422164 | 0.04 |
ENSRNOT00000017174
|
Nqo1
|
NAD(P)H quinone dehydrogenase 1 |
| chr1_+_91042635 | 0.04 |
ENSRNOT00000028211
|
LOC103690005
|
tubulin-folding cofactor B |
| chrX_-_72078551 | 0.04 |
ENSRNOT00000076978
|
Rps4x
|
ribosomal protein S4, X-linked |
| chrX_-_77418525 | 0.04 |
ENSRNOT00000092303
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr10_+_57239993 | 0.04 |
ENSRNOT00000067919
|
LOC687707
|
hypothetical protein LOC687707 |
| chr1_+_84584596 | 0.04 |
ENSRNOT00000032933
|
LOC100910046
|
zinc finger protein 60-like |
| chr1_-_38538987 | 0.04 |
ENSRNOT00000090406
ENSRNOT00000071275 |
LOC102551340
|
zinc finger protein 728-like |
| chr8_+_12355767 | 0.04 |
ENSRNOT00000068445
|
Fam76b
|
family with sequence similarity 76, member B |
| chr7_+_77678968 | 0.04 |
ENSRNOT00000006917
|
Atp6v1c1
|
ATPase H+ transporting V1 subunit C1 |
| chr1_-_214159157 | 0.04 |
ENSRNOT00000091292
ENSRNOT00000022241 |
Rnh1
|
ribonuclease/angiogenin inhibitor 1 |
| chr10_-_41408518 | 0.04 |
ENSRNOT00000018967
|
Nmur2
|
neuromedin U receptor 2 |
| chr15_+_31057655 | 0.04 |
ENSRNOT00000073618
|
AABR07017783.2
|
|
| chr2_-_182846061 | 0.04 |
ENSRNOT00000013025
|
Tlr2
|
toll-like receptor 2 |
| chr17_+_29639079 | 0.04 |
ENSRNOT00000021877
|
Rpp40
|
ribonuclease P/MRP 40 subunit |
| chr14_+_18983853 | 0.04 |
ENSRNOT00000003836
|
Rassf6
|
Ras association domain family member 6 |
| chr15_+_30612409 | 0.04 |
ENSRNOT00000072977
|
AABR07017748.1
|
|
| chr10_-_16689321 | 0.04 |
ENSRNOT00000028173
|
Bnip1
|
BCL2/adenovirus E1B interacting protein 1 |
| chr14_+_83560541 | 0.04 |
ENSRNOT00000057738
ENSRNOT00000085228 |
Pik3ip1
|
phosphoinositide-3-kinase interacting protein 1 |
| chr11_-_75144903 | 0.04 |
ENSRNOT00000002329
|
Hrasls
|
HRAS-like suppressor |
| chr15_-_8914501 | 0.04 |
ENSRNOT00000008752
|
Thrb
|
thyroid hormone receptor beta |
| chr4_-_84127331 | 0.04 |
ENSRNOT00000085812
|
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chr18_+_29960072 | 0.04 |
ENSRNOT00000071366
|
AC103179.1
|
|
| chr3_-_133131192 | 0.04 |
ENSRNOT00000055630
|
Tasp1
|
taspase 1 |
| chr2_-_195423787 | 0.04 |
ENSRNOT00000071603
|
LOC103689947
|
selenium-binding protein 1 |
| chr15_-_35417273 | 0.04 |
ENSRNOT00000083961
ENSRNOT00000041430 |
Gzmb
|
granzyme B |
| chr2_+_187288644 | 0.04 |
ENSRNOT00000035271
|
mrpl24
|
mitochondrial ribosomal protein L24 |
| chrX_-_83788325 | 0.04 |
ENSRNOT00000037149
|
Satl1
|
spermidine/spermine N1-acetyl transferase-like 1 |
| chrX_+_16050780 | 0.04 |
ENSRNOT00000079054
|
Clcn5
|
chloride voltage-gated channel 5 |
| chr4_-_131694755 | 0.04 |
ENSRNOT00000013271
|
Foxp1
|
forkhead box P1 |
| chr10_-_56276764 | 0.04 |
ENSRNOT00000049048
|
Eif4a1
|
eukaryotic translation initiation factor 4A1 |
| chr8_-_17524839 | 0.04 |
ENSRNOT00000081679
|
Naalad2
|
N-acetylated alpha-linked acidic dipeptidase 2 |
| chrX_-_156891213 | 0.04 |
ENSRNOT00000082899
|
Avpr2
|
arginine vasopressin receptor 2 |
| chr20_-_27682861 | 0.04 |
ENSRNOT00000057317
|
Fam26f
|
family with sequence similarity 26, member F |
| chr11_+_27004320 | 0.04 |
ENSRNOT00000002182
|
N6amt1
|
N-6 adenine-specific DNA methyltransferase 1 (putative) |
| chr17_-_70548071 | 0.04 |
ENSRNOT00000073144
|
Il2ra
|
interleukin 2 receptor subunit alpha |
| chr20_-_45260119 | 0.04 |
ENSRNOT00000000718
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr5_-_147303346 | 0.04 |
ENSRNOT00000009153
|
Hpca
|
hippocalcin |
| chrX_-_77529374 | 0.04 |
ENSRNOT00000078213
|
Fndc3c1
|
fibronectin type III domain containing 3C1 |
| chr1_+_97712177 | 0.03 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chr6_+_86823684 | 0.03 |
ENSRNOT00000086081
ENSRNOT00000006574 |
Fancm
|
Fanconi anemia, complementation group M |
| chr11_-_88092756 | 0.03 |
ENSRNOT00000002542
|
Ydjc
|
YdjC homolog (bacterial) |
| chr5_-_107858104 | 0.03 |
ENSRNOT00000092196
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr8_+_63379087 | 0.03 |
ENSRNOT00000012091
ENSRNOT00000012295 |
Nptn
|
neuroplastin |
| chr12_-_37682964 | 0.03 |
ENSRNOT00000001421
|
RGD1563482
|
similar to hypothetical protein FLJ38663 |
| chr9_+_101284902 | 0.03 |
ENSRNOT00000065533
|
NEWGENE_1308624
|
sialidase 4 |
| chr6_+_107156263 | 0.03 |
ENSRNOT00000003834
|
Rbm25
|
RNA binding motif protein 25 |
| chrX_-_16537888 | 0.03 |
ENSRNOT00000060247
|
Dgkk
|
diacylglycerol kinase kappa |
| chr10_+_14656812 | 0.03 |
ENSRNOT00000024904
|
Prss34
|
protease, serine, 34 |
| chr15_+_33885106 | 0.03 |
ENSRNOT00000024483
|
Dhrs2
|
dehydrogenase/reductase 2 |
| chr1_-_91042230 | 0.03 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chr11_-_61470046 | 0.03 |
ENSRNOT00000073436
|
LOC102553099
|
N-alpha-acetyltransferase 50-like |
| chr3_-_110828879 | 0.03 |
ENSRNOT00000013963
|
Ccdc32
|
coiled-coil domain containing 32 |
| chr3_+_56966654 | 0.03 |
ENSRNOT00000088097
|
Gorasp2
|
golgi reassembly stacking protein 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Six6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.1 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.0 | 0.1 | GO:1905133 | metaphase/anaphase transition of meiotic cell cycle(GO:0044785) regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902102) negative regulation of metaphase/anaphase transition of meiotic cell cycle(GO:1902103) regulation of meiotic chromosome separation(GO:1905132) negative regulation of meiotic chromosome separation(GO:1905133) |
| 0.0 | 0.1 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.0 | 0.1 | GO:2000532 | cellular response to vitamin K(GO:0071307) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) negative regulation of dendritic cell apoptotic process(GO:2000669) |
| 0.0 | 0.1 | GO:0019389 | glucuronoside metabolic process(GO:0019389) response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 0.0 | 0.2 | GO:0048478 | replication fork protection(GO:0048478) |
| 0.0 | 0.1 | GO:0016480 | negative regulation of transcription from RNA polymerase III promoter(GO:0016480) |
| 0.0 | 0.2 | GO:0034124 | regulation of MyD88-dependent toll-like receptor signaling pathway(GO:0034124) |
| 0.0 | 0.3 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.2 | GO:0050427 | flavonoid metabolic process(GO:0009812) purine ribonucleoside bisphosphate metabolic process(GO:0034035) 3'-phosphoadenosine 5'-phosphosulfate metabolic process(GO:0050427) |
| 0.0 | 0.1 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.1 | GO:0046391 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:0038110 | interleukin-2-mediated signaling pathway(GO:0038110) |
| 0.0 | 0.0 | GO:0060460 | left lung morphogenesis(GO:0060460) |
| 0.0 | 0.0 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.1 | GO:0048687 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.1 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.0 | 0.0 | GO:0032430 | positive regulation of phospholipase A2 activity(GO:0032430) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) central nervous system myelin formation(GO:0032289) positive regulation of interleukin-18 production(GO:0032741) detection of diacyl bacterial lipopeptide(GO:0042496) response to immune response of other organism involved in symbiotic interaction(GO:0052564) response to host immune response(GO:0052572) detection of bacterial lipopeptide(GO:0070340) |
| 0.0 | 0.0 | GO:0060545 | positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.0 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:1902965 | regulation of protein localization to early endosome(GO:1902965) positive regulation of protein localization to early endosome(GO:1902966) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.0 | GO:0035355 | Toll-like receptor 2-Toll-like receptor 6 protein complex(GO:0035355) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0061617 | MICOS complex(GO:0061617) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0050294 | aryl sulfotransferase activity(GO:0004062) steroid sulfotransferase activity(GO:0050294) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.2 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0004911 | interleukin-2 receptor activity(GO:0004911) |
| 0.0 | 0.1 | GO:0034437 | glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.0 | GO:0042498 | diacyl lipopeptide binding(GO:0042498) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.0 | GO:0019809 | spermidine binding(GO:0019809) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0097493 | structural molecule activity conferring elasticity(GO:0097493) |
| 0.0 | 0.0 | GO:0055100 | adiponectin binding(GO:0055100) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.2 | GO:0042288 | MHC class I protein binding(GO:0042288) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.0 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.0 | 0.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |