Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
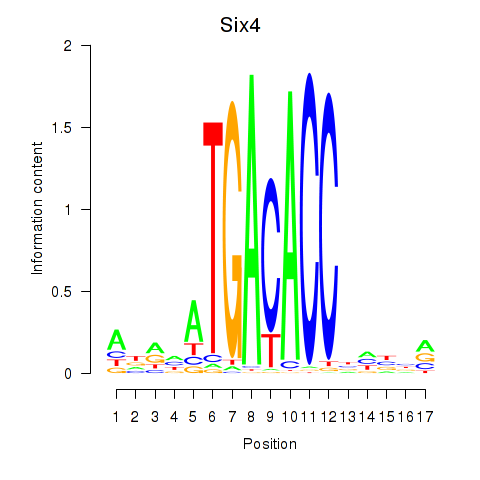
Results for Six4
Z-value: 0.27
Transcription factors associated with Six4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Six4
|
ENSRNOG00000007250 | SIX homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six4 | rn6_v1_chr6_-_95998529_95998529 | -0.16 | 8.0e-01 | Click! |
Activity profile of Six4 motif
Sorted Z-values of Six4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_+_120580743 | 0.15 |
ENSRNOT00000017181
|
Maff
|
MAF bZIP transcription factor F |
| chr8_-_46694613 | 0.12 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr1_-_218657925 | 0.08 |
ENSRNOT00000020425
|
Gal
|
galanin and GMAP prepropeptide |
| chr4_+_33638709 | 0.07 |
ENSRNOT00000009888
ENSRNOT00000034719 ENSRNOT00000052333 |
Tac1
|
tachykinin, precursor 1 |
| chr1_+_80135391 | 0.07 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chr7_+_72599479 | 0.06 |
ENSRNOT00000018963
|
AABR07057460.1
|
|
| chr1_+_221773254 | 0.06 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr12_+_27155587 | 0.06 |
ENSRNOT00000044800
|
AABR07035916.1
|
|
| chr1_-_166037424 | 0.05 |
ENSRNOT00000026115
|
P2ry2
|
purinergic receptor P2Y2 |
| chr15_+_33755478 | 0.04 |
ENSRNOT00000034073
|
Thtpa
|
thiamine triphosphatase |
| chr5_-_167030441 | 0.03 |
ENSRNOT00000023543
|
H6pd
|
hexose-6-phosphate dehydrogenase (glucose 1-dehydrogenase) |
| chr8_-_111965889 | 0.03 |
ENSRNOT00000032376
|
Bfsp2
|
beaded filament structural protein 2 |
| chr1_-_209641123 | 0.03 |
ENSRNOT00000021702
|
Ebf3
|
early B-cell factor 3 |
| chr20_+_5040337 | 0.03 |
ENSRNOT00000068435
|
Clic1
|
chloride intracellular channel 1 |
| chr9_-_81565416 | 0.03 |
ENSRNOT00000083582
|
Aamp
|
angio-associated, migratory cell protein |
| chr2_-_26699333 | 0.02 |
ENSRNOT00000024459
|
Sv2c
|
synaptic vesicle glycoprotein 2c |
| chr16_-_79671719 | 0.02 |
ENSRNOT00000015908
|
Myom2
|
myomesin 2 |
| chr4_+_25507463 | 0.02 |
ENSRNOT00000007977
|
Steap2
|
STEAP2 metalloreductase |
| chr15_+_24254042 | 0.02 |
ENSRNOT00000092161
|
Fbxo34
|
F-box protein 34 |
| chr15_+_24267323 | 0.02 |
ENSRNOT00000015778
|
Fbxo34
|
F-box protein 34 |
| chr9_+_60883981 | 0.02 |
ENSRNOT00000081002
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr7_-_144880092 | 0.02 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
| chr18_+_38847632 | 0.02 |
ENSRNOT00000014916
|
LOC102554034
|
ELMO domain-containing protein 2-like |
| chr5_+_138354324 | 0.02 |
ENSRNOT00000066742
|
Gm17728
|
predicted gene, 17728 |
| chr1_-_141893674 | 0.02 |
ENSRNOT00000019059
ENSRNOT00000085988 |
Idh2
|
isocitrate dehydrogenase (NADP(+)) 2, mitochondrial |
| chr7_+_11660934 | 0.02 |
ENSRNOT00000022336
|
Lmnb2
|
lamin B2 |
| chr17_+_89171250 | 0.01 |
ENSRNOT00000024901
|
Gad2
|
glutamate decarboxylase 2 |
| chr17_-_15566332 | 0.01 |
ENSRNOT00000093743
|
Ecm2
|
extracellular matrix protein 2 |
| chrX_-_111887906 | 0.01 |
ENSRNOT00000085118
|
Tsc22d3
|
TSC22 domain family, member 3 |
| chr17_+_81798756 | 0.01 |
ENSRNOT00000066826
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr10_+_88326337 | 0.01 |
ENSRNOT00000022029
ENSRNOT00000076461 |
Fkbp10
|
FK506 binding protein 10 |
| chr9_+_17728816 | 0.01 |
ENSRNOT00000065754
|
Capn11
|
calpain 11 |
| chr1_-_222235102 | 0.01 |
ENSRNOT00000028725
|
Fkbp2
|
FK506 binding protein 2 |
| chr5_-_12526962 | 0.01 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr1_+_177249226 | 0.01 |
ENSRNOT00000021671
|
Parva
|
parvin, alpha |
| chr1_-_190596431 | 0.01 |
ENSRNOT00000079335
|
Pdzd9
|
PDZ domain containing 9 |
| chr1_+_99616447 | 0.01 |
ENSRNOT00000029197
|
Klk14
|
kallikrein related-peptidase 14 |
| chr14_-_21709084 | 0.01 |
ENSRNOT00000087477
|
Smr3b
|
submaxillary gland androgen regulated protein 3B |
| chr16_+_20740826 | 0.01 |
ENSRNOT00000038057
|
Crtc1
|
CREB regulated transcription coactivator 1 |
| chrX_+_112020646 | 0.01 |
ENSRNOT00000079841
|
Mid2
|
midline 2 |
| chr1_+_70260041 | 0.01 |
ENSRNOT00000020416
|
Zim1
|
zinc finger, imprinted 1 |
| chr4_-_153465203 | 0.01 |
ENSRNOT00000016776
|
Bid
|
BH3 interacting domain death agonist |
| chr16_-_19366485 | 0.01 |
ENSRNOT00000077789
|
Rab8a
|
RAB8A, member RAS oncogene family |
| chr8_+_117355943 | 0.01 |
ENSRNOT00000027318
|
Dalrd3
|
DALR anticodon binding domain containing 3 |
| chr10_+_75149814 | 0.01 |
ENSRNOT00000011500
|
Mks1
|
Meckel syndrome, type 1 |
| chr6_+_48708368 | 0.01 |
ENSRNOT00000005905
|
Myt1l
|
myelin transcription factor 1-like |
| chr1_-_53038229 | 0.01 |
ENSRNOT00000017282
|
Mpc1
|
mitochondrial pyruvate carrier 1 |
| chr1_-_229601032 | 0.01 |
ENSRNOT00000016690
|
Cntf
|
ciliary neurotrophic factor |
| chr20_+_11863082 | 0.01 |
ENSRNOT00000001640
|
Fam207a
|
family with sequence similarity 207, member A |
| chr8_+_32165810 | 0.01 |
ENSRNOT00000007539
|
Zbtb44
|
zinc finger and BTB domain containing 44 |
| chr10_-_40381886 | 0.01 |
ENSRNOT00000050213
|
LOC100912427
|
nuclease-sensitive element-binding protein 1-like |
| chr5_-_152247332 | 0.01 |
ENSRNOT00000077165
|
Lin28a
|
lin-28 homolog A |
| chr12_-_50400765 | 0.01 |
ENSRNOT00000071613
|
Crybb1
|
crystallin, beta B1 |
| chr6_-_41870046 | 0.01 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr1_-_99135977 | 0.00 |
ENSRNOT00000056515
|
Vom2r38
|
vomeronasal 2 receptor, 38 |
| chr1_-_73121085 | 0.00 |
ENSRNOT00000073464
|
LOC684545
|
similar to NACHT, leucine rich repeat and PYD containing 2 |
| chr10_-_18579632 | 0.00 |
ENSRNOT00000040737
|
Kcnip1
|
potassium voltage-gated channel interacting protein 1 |
| chr1_-_199395363 | 0.00 |
ENSRNOT00000090368
ENSRNOT00000026587 |
Prss36
|
protease, serine, 36 |
| chr11_-_61530567 | 0.00 |
ENSRNOT00000076277
|
Naa50
|
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr15_+_5319916 | 0.00 |
ENSRNOT00000046644
|
LOC102546495
|
disks large homolog 5-like |
| chr10_-_104624757 | 0.00 |
ENSRNOT00000087759
|
Unc13d
|
unc-13 homolog D |
| chr14_+_77079402 | 0.00 |
ENSRNOT00000042200
|
Slc2a9
|
solute carrier family 2 member 9 |
| chr11_+_20474483 | 0.00 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr9_-_75528644 | 0.00 |
ENSRNOT00000019283
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chrX_+_16050780 | 0.00 |
ENSRNOT00000079054
|
Clcn5
|
chloride voltage-gated channel 5 |
| chr3_+_112916217 | 0.00 |
ENSRNOT00000030065
|
Tmem62
|
transmembrane protein 62 |
| chr8_-_77398156 | 0.00 |
ENSRNOT00000091858
ENSRNOT00000085349 ENSRNOT00000082763 |
Lipc
|
lipase C, hepatic type |
Network of associatons between targets according to the STRING database.
First level regulatory network of Six4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:2000851 | positive regulation of glucocorticoid secretion(GO:2000851) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.0 | GO:0006772 | thiamine metabolic process(GO:0006772) thiamine diphosphate metabolic process(GO:0042357) thiamine-containing compound metabolic process(GO:0042723) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071553 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0004966 | galanin receptor activity(GO:0004966) |