Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Six3_Six1_Six2
Z-value: 0.29
Transcription factors associated with Six3_Six1_Six2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
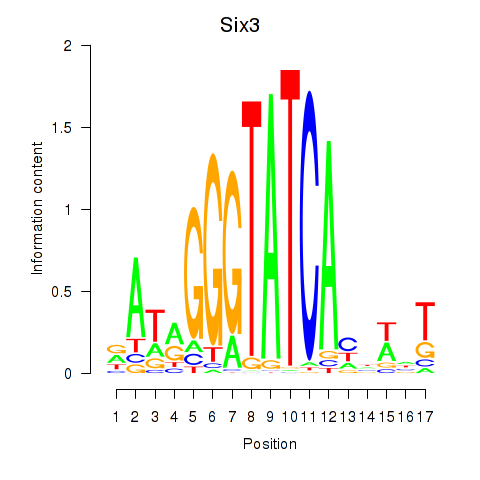
Six3
|
ENSRNOG00000057031 | SIX homeobox 3 |
|
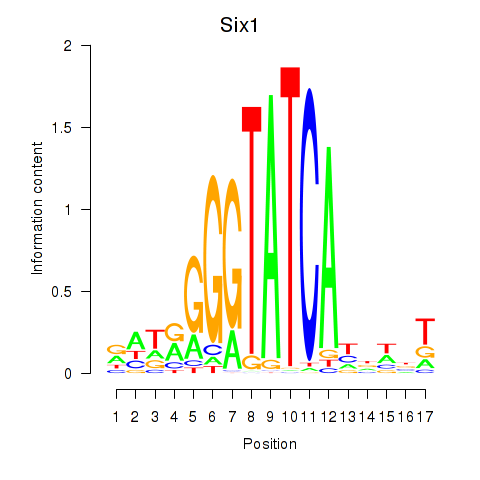
Six1
|
ENSRNOG00000022777 | SIX homeobox 1 |
|
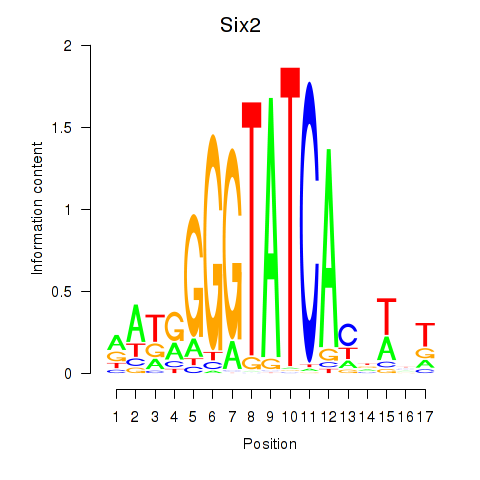
Six2
|
ENSRNOG00000058827 | SIX homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Six2 | rn6_v1_chr6_-_8956276_8956276 | 0.49 | 4.0e-01 | Click! |
| Six1 | rn6_v1_chr6_-_95934296_95934296 | -0.29 | 6.3e-01 | Click! |
| Six3 | rn6_v1_chr6_+_8886591_8886730 | -0.12 | 8.5e-01 | Click! |
Activity profile of Six3_Six1_Six2 motif
Sorted Z-values of Six3_Six1_Six2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_111948753 | 0.24 |
ENSRNOT00000048074
|
Hsd11b1
|
hydroxysteroid 11-beta dehydrogenase 1 |
| chr6_+_64224861 | 0.22 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chrX_+_19854836 | 0.20 |
ENSRNOT00000075010
|
AABR07037395.2
|
|
| chr5_+_173148884 | 0.16 |
ENSRNOT00000041753
|
LOC100364191
|
hCG1994130-like |
| chr10_+_14492844 | 0.14 |
ENSRNOT00000023615
|
Clcn7
|
chloride voltage-gated channel 7 |
| chrX_-_114232939 | 0.13 |
ENSRNOT00000042639
|
Ammecr1
|
Alport syndrome, mental retardation, midface hypoplasia and elliptocytosis chromosomal region gene 1 |
| chr8_-_76940094 | 0.10 |
ENSRNOT00000082709
ENSRNOT00000084313 |
Rnf111
|
ring finger protein 111 |
| chr10_-_36322356 | 0.09 |
ENSRNOT00000000248
|
Zfp354c
|
zinc finger protein 354C |
| chr3_-_93789617 | 0.08 |
ENSRNOT00000078616
|
Caprin1
|
cell cycle associated protein 1 |
| chr16_-_19544727 | 0.08 |
ENSRNOT00000074514
|
Zfp617
|
zinc finger protein 617 |
| chr6_+_86823684 | 0.07 |
ENSRNOT00000086081
ENSRNOT00000006574 |
Fancm
|
Fanconi anemia, complementation group M |
| chr5_+_124442293 | 0.07 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr5_+_9230984 | 0.06 |
ENSRNOT00000009136
|
Vcpip1
|
valosin containing protein interacting protein 1 |
| chr2_-_195935878 | 0.06 |
ENSRNOT00000028440
|
Cgn
|
cingulin |
| chr1_+_201981357 | 0.05 |
ENSRNOT00000027999
|
Acadsb
|
acyl-CoA dehydrogenase, short/branched chain |
| chr10_+_71546977 | 0.05 |
ENSRNOT00000041140
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr1_-_198104109 | 0.05 |
ENSRNOT00000026186
|
Sult1a1
|
sulfotransferase family 1A member 1 |
| chr2_-_187786700 | 0.04 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr3_-_158328881 | 0.04 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr2_-_80667481 | 0.04 |
ENSRNOT00000016784
|
Trio
|
trio Rho guanine nucleotide exchange factor |
| chr1_+_57692836 | 0.04 |
ENSRNOT00000083968
ENSRNOT00000019358 |
Chd1
|
chromodomain helicase DNA binding protein 1 |
| chr4_+_170518673 | 0.04 |
ENSRNOT00000011803
|
Atf7ip
|
activating transcription factor 7 interacting protein |
| chr4_-_150520363 | 0.04 |
ENSRNOT00000065579
|
Zfp9
|
zinc finger protein 9 |
| chr6_+_96834525 | 0.04 |
ENSRNOT00000077935
|
Hif1a
|
hypoxia inducible factor 1 alpha subunit |
| chr8_+_115546712 | 0.03 |
ENSRNOT00000079801
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chrX_+_156336450 | 0.03 |
ENSRNOT00000082124
|
Slc10a3
|
solute carrier family 10, member 3 |
| chr9_-_7891514 | 0.03 |
ENSRNOT00000072684
|
Pot1b
|
protection of telomeres 1B |
| chr10_+_70298519 | 0.03 |
ENSRNOT00000035730
ENSRNOT00000076233 ENSRNOT00000076445 ENSRNOT00000076133 |
Slfn5
|
schlafen family member 5 |
| chr3_-_145641654 | 0.03 |
ENSRNOT00000008101
ENSRNOT00000008047 |
LOC102554315
|
zinc finger protein 596-like |
| chr11_+_86903122 | 0.03 |
ENSRNOT00000048063
|
Zdhhc8
|
zinc finger, DHHC-type containing 8 |
| chr7_-_60860990 | 0.03 |
ENSRNOT00000009511
|
Rap1b
|
RAP1B, member of RAS oncogene family |
| chr1_-_201981229 | 0.02 |
ENSRNOT00000077246
ENSRNOT00000035531 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr9_-_11108741 | 0.02 |
ENSRNOT00000072357
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr10_+_62566732 | 0.02 |
ENSRNOT00000021368
|
Taok1
|
TAO kinase 1 |
| chr8_-_80631873 | 0.02 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr7_+_80750725 | 0.02 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chr18_+_30856847 | 0.02 |
ENSRNOT00000027019
|
AABR07031734.1
|
|
| chr11_-_11585078 | 0.02 |
ENSRNOT00000088878
|
Robo2
|
roundabout guidance receptor 2 |
| chr16_-_81714346 | 0.02 |
ENSRNOT00000092552
|
Lamp1
|
lysosomal-associated membrane protein 1 |
| chrX_-_56765893 | 0.02 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr7_+_117643206 | 0.02 |
ENSRNOT00000077588
|
Adck5
|
aarF domain containing kinase 5 |
| chr2_-_124047044 | 0.02 |
ENSRNOT00000065384
|
Cetn4
|
centrin 4 |
| chr10_+_53713938 | 0.02 |
ENSRNOT00000004236
ENSRNOT00000086599 ENSRNOT00000085582 |
Myh2
|
myosin heavy chain 2 |
| chr1_-_189182306 | 0.02 |
ENSRNOT00000021249
|
Gp2
|
glycoprotein 2 |
| chr2_-_26011429 | 0.02 |
ENSRNOT00000065143
|
Arhgef28
|
Rho guanine nucleotide exchange factor 28 |
| chr13_+_56546021 | 0.02 |
ENSRNOT00000016797
|
Aspm
|
abnormal spindle microtubule assembly |
| chr1_+_225899313 | 0.02 |
ENSRNOT00000031612
|
Scgb2a1
|
secretoglobin, family 2A, member 1 |
| chr12_+_17253791 | 0.02 |
ENSRNOT00000083814
|
Zfand2a
|
zinc finger AN1-type containing 2A |
| chr1_-_213650247 | 0.02 |
ENSRNOT00000019679
|
Cox8b
|
cytochrome c oxidase, subunit VIIIb |
| chrX_+_105575759 | 0.02 |
ENSRNOT00000088760
|
Armcx3
|
armadillo repeat containing, X-linked 3 |
| chr10_-_13892997 | 0.02 |
ENSRNOT00000004192
|
Traf7
|
TNF receptor associated factor 7 |
| chr7_+_30699476 | 0.02 |
ENSRNOT00000059269
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr13_-_34253910 | 0.02 |
ENSRNOT00000092847
|
Tsn
|
translin |
| chr2_-_247446882 | 0.02 |
ENSRNOT00000021963
|
Bmpr1b
|
bone morphogenetic protein receptor type 1B |
| chr1_-_100530183 | 0.01 |
ENSRNOT00000067754
|
Mybpc2
|
myosin binding protein C, fast-type |
| chr1_+_87045796 | 0.01 |
ENSRNOT00000027620
|
Lgals7
|
galectin 7 |
| chr2_-_182846061 | 0.01 |
ENSRNOT00000013025
|
Tlr2
|
toll-like receptor 2 |
| chr6_+_48452369 | 0.01 |
ENSRNOT00000044310
|
Myt1l
|
myelin transcription factor 1-like |
| chr12_-_16201632 | 0.01 |
ENSRNOT00000001688
|
Chst12
|
carbohydrate sulfotransferase 12 |
| chr11_+_73936750 | 0.01 |
ENSRNOT00000002350
|
Atp13a3
|
ATPase 13A3 |
| chr17_-_67945037 | 0.01 |
ENSRNOT00000023033
|
Klf6
|
Kruppel-like factor 6 |
| chr17_+_77195247 | 0.01 |
ENSRNOT00000081940
|
Optn
|
optineurin |
| chr8_+_133210473 | 0.01 |
ENSRNOT00000082774
|
Ccr5
|
chemokine (C-C motif) receptor 5 |
| chr14_+_48726045 | 0.01 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr6_-_115352681 | 0.01 |
ENSRNOT00000005873
|
Gtf2a1
|
general transcription factor 2A subunit 1 |
| chr8_-_21481735 | 0.01 |
ENSRNOT00000038069
|
Zfp426
|
zinc finger protein 426 |
| chr5_+_146294030 | 0.01 |
ENSRNOT00000073637
|
AABR07049960.1
|
|
| chr1_-_167884690 | 0.01 |
ENSRNOT00000091372
|
Olr61
|
olfactory receptor 61 |
| chr3_+_119173818 | 0.01 |
ENSRNOT00000015124
|
Usp8
|
ubiquitin specific peptidase 8 |
| chr8_-_21453190 | 0.01 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chr17_-_84249243 | 0.01 |
ENSRNOT00000082549
|
Nebl
|
nebulette |
| chr13_+_113373578 | 0.01 |
ENSRNOT00000009900
|
Plxna2
|
plexin A2 |
| chr1_-_167700332 | 0.01 |
ENSRNOT00000092890
|
Trim21
|
tripartite motif-containing 21 |
| chr4_+_56748494 | 0.01 |
ENSRNOT00000065747
|
LOC103692087
|
uncharacterized LOC103692087 |
| chr1_+_87045335 | 0.01 |
ENSRNOT00000084393
|
Lgals7
|
galectin 7 |
| chr5_+_158090173 | 0.01 |
ENSRNOT00000088766
ENSRNOT00000079516 ENSRNOT00000092026 |
Tas1r2
|
taste 1 receptor member 2 |
| chr3_-_113091770 | 0.01 |
ENSRNOT00000068132
|
Lcmt2
|
leucine carboxyl methyltransferase 2 |
| chr1_-_189181901 | 0.01 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr9_-_92524739 | 0.01 |
ENSRNOT00000089889
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr20_-_4818568 | 0.01 |
ENSRNOT00000001115
|
Ddx39b
|
DExD-box helicase 39B |
| chr8_+_115546893 | 0.01 |
ENSRNOT00000018932
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr1_-_77893509 | 0.01 |
ENSRNOT00000015059
|
Gltscr1
|
glioma tumor suppressor candidate region gene 1 |
| chr2_-_211690716 | 0.01 |
ENSRNOT00000027690
|
Fndc7
|
fibronectin type III domain containing 7 |
| chrX_+_9956370 | 0.01 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr7_-_30274984 | 0.01 |
ENSRNOT00000092527
ENSRNOT00000010207 |
Slc17a8
|
solute carrier family 17 member 8 |
| chr4_-_115015965 | 0.01 |
ENSRNOT00000014603
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr1_-_226255886 | 0.01 |
ENSRNOT00000027842
|
Fen1
|
flap structure-specific endonuclease 1 |
| chr19_+_15195565 | 0.00 |
ENSRNOT00000090865
ENSRNOT00000078874 |
Ces1d
|
carboxylesterase 1D |
| chr8_+_69753373 | 0.00 |
ENSRNOT00000066973
|
Tipin
|
timeless interacting protein |
| chr7_-_10430992 | 0.00 |
ENSRNOT00000047433
|
Zfp709l1
|
zinc finger protein 709-like 1 |
| chr14_-_25585222 | 0.00 |
ENSRNOT00000042106
|
RGD1565048
|
similar to 60S ribosomal protein L9 |
| chr2_-_123851854 | 0.00 |
ENSRNOT00000023327
|
Il2
|
interleukin 2 |
| chr8_+_91368111 | 0.00 |
ENSRNOT00000083151
|
Ttk
|
Ttk protein kinase |
| chrX_+_77076106 | 0.00 |
ENSRNOT00000091527
ENSRNOT00000089381 |
Atp7a
|
ATPase copper transporting alpha |
| chr2_+_83393282 | 0.00 |
ENSRNOT00000044871
|
Ctnnd2
|
catenin delta 2 |
| chr3_-_110828879 | 0.00 |
ENSRNOT00000013963
|
Ccdc32
|
coiled-coil domain containing 32 |
| chr11_+_73693814 | 0.00 |
ENSRNOT00000081081
ENSRNOT00000002354 ENSRNOT00000090940 |
Lsg1
|
large 60S subunit nuclear export GTPase 1 |
| chr18_-_27452420 | 0.00 |
ENSRNOT00000037888
|
Cdc23
|
cell division cycle 23 |
| chr1_-_212622537 | 0.00 |
ENSRNOT00000025609
|
Sprn
|
shadow of prion protein homolog (zebrafish) |
| chr10_+_31813814 | 0.00 |
ENSRNOT00000009573
|
Havcr1
|
hepatitis A virus cellular receptor 1 |
| chr8_-_46694613 | 0.00 |
ENSRNOT00000084514
|
AC133265.1
|
|
| chr19_+_38422164 | 0.00 |
ENSRNOT00000017174
|
Nqo1
|
NAD(P)H quinone dehydrogenase 1 |
| chr4_+_72718458 | 0.00 |
ENSRNOT00000044780
|
Arhgef5
|
Rho guanine nucleotide exchange factor 5 |
| chr13_-_78521911 | 0.00 |
ENSRNOT00000087506
|
Rabgap1l
|
RAB GTPase activating protein 1-like |
| chr18_+_86071662 | 0.00 |
ENSRNOT00000064901
|
Rttn
|
rotatin |
| chr2_+_26240385 | 0.00 |
ENSRNOT00000024292
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr4_-_176528110 | 0.00 |
ENSRNOT00000049569
|
Slco1a2
|
solute carrier organic anion transporter family, member 1A2 |
| chr15_-_14737704 | 0.00 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
Network of associatons between targets according to the STRING database.
First level regulatory network of Six3_Six1_Six2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006713 | glucocorticoid catabolic process(GO:0006713) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.0 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.0 | GO:0071250 | cellular response to carbon monoxide(GO:0071245) cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.0 | GO:0050656 | aryl sulfotransferase activity(GO:0004062) steroid sulfotransferase activity(GO:0050294) 3'-phosphoadenosine 5'-phosphosulfate binding(GO:0050656) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |