Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Scrt2
Z-value: 0.42
Transcription factors associated with Scrt2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Scrt2
|
ENSRNOG00000005148 | scratch family transcriptional repressor 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Scrt2 | rn6_v1_chr3_+_147585947_147585947 | -0.26 | 6.7e-01 | Click! |
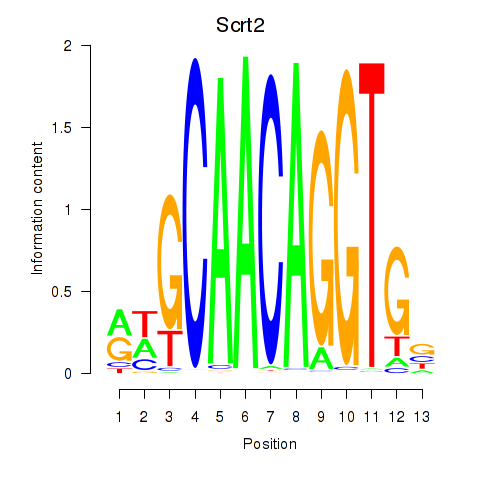
Activity profile of Scrt2 motif
Sorted Z-values of Scrt2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_49676540 | 0.18 |
ENSRNOT00000022032
ENSRNOT00000082205 |
Fxyd6
|
FXYD domain-containing ion transport regulator 6 |
| chr1_+_53220397 | 0.13 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr16_-_19942343 | 0.12 |
ENSRNOT00000087162
ENSRNOT00000091906 |
Bst2
|
bone marrow stromal cell antigen 2 |
| chr1_-_15301998 | 0.12 |
ENSRNOT00000016400
|
Slc35d3
|
solute carrier family 35, member D3 |
| chr1_+_78820262 | 0.11 |
ENSRNOT00000022441
|
Gng8
|
G protein subunit gamma 8 |
| chr2_+_121165137 | 0.09 |
ENSRNOT00000016236
|
Sox2
|
SRY box 2 |
| chr20_-_47306318 | 0.09 |
ENSRNOT00000075151
|
Nr2e1
|
nuclear receptor subfamily 2, group E, member 1 |
| chr15_+_67555835 | 0.09 |
ENSRNOT00000045882
|
Pcdh17
|
protocadherin 17 |
| chr3_+_100768637 | 0.09 |
ENSRNOT00000083542
|
Bdnf
|
brain-derived neurotrophic factor |
| chr12_+_2534212 | 0.08 |
ENSRNOT00000001399
|
Ctxn1
|
cortexin 1 |
| chr1_-_188101566 | 0.08 |
ENSRNOT00000092435
|
Syt17
|
synaptotagmin 17 |
| chr1_+_242959488 | 0.08 |
ENSRNOT00000015668
|
Dock8
|
dedicator of cytokinesis 8 |
| chr1_+_205842489 | 0.08 |
ENSRNOT00000081610
|
Fank1
|
fibronectin type 3 and ankyrin repeat domains 1 |
| chr10_+_3225481 | 0.07 |
ENSRNOT00000093598
|
Ntan1
|
N-terminal asparagine amidase |
| chr4_-_146839397 | 0.07 |
ENSRNOT00000010338
|
Vgll4
|
vestigial-like family member 4 |
| chr2_+_28049217 | 0.07 |
ENSRNOT00000022144
|
Enc1
|
ectodermal-neural cortex 1 |
| chr2_+_209766512 | 0.07 |
ENSRNOT00000092240
|
Kcna3
|
potassium voltage-gated channel subfamily A member 3 |
| chrX_-_123254557 | 0.06 |
ENSRNOT00000039809
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr1_+_106896790 | 0.06 |
ENSRNOT00000042952
ENSRNOT00000064193 |
Ano5
|
anoctamin 5 |
| chr2_-_210874304 | 0.06 |
ENSRNOT00000088657
|
Ampd2
|
adenosine monophosphate deaminase 2 |
| chr1_+_167320797 | 0.06 |
ENSRNOT00000076923
|
Pgap2
|
post-GPI attachment to proteins 2 |
| chr13_+_101698096 | 0.06 |
ENSRNOT00000089309
|
Aida
|
axin interactor, dorsalization associated |
| chr6_+_86785771 | 0.06 |
ENSRNOT00000066702
|
Prpf39
|
pre-mRNA processing factor 39 |
| chr10_-_13898855 | 0.06 |
ENSRNOT00000004249
|
Rab26
|
RAB26, member RAS oncogene family |
| chr7_-_59514939 | 0.06 |
ENSRNOT00000085579
|
Kcnmb4
|
potassium calcium-activated channel subfamily M regulatory beta subunit 4 |
| chr1_-_199624783 | 0.06 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
| chr20_+_37876650 | 0.06 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr14_-_43143973 | 0.05 |
ENSRNOT00000003248
|
Uchl1
|
ubiquitin C-terminal hydrolase L1 |
| chrX_-_70978952 | 0.05 |
ENSRNOT00000076910
|
Slc7a3
|
solute carrier family 7 member 3 |
| chr17_+_57074525 | 0.05 |
ENSRNOT00000020012
ENSRNOT00000074146 |
Crem
|
cAMP responsive element modulator |
| chr20_+_5050327 | 0.05 |
ENSRNOT00000083353
|
Ddah2
|
dimethylarginine dimethylaminohydrolase 2 |
| chr7_+_12974169 | 0.05 |
ENSRNOT00000010555
|
C2cd4c
|
C2 calcium-dependent domain containing 4C |
| chrX_-_70978714 | 0.05 |
ENSRNOT00000076336
|
Slc7a3
|
solute carrier family 7 member 3 |
| chr3_-_113405829 | 0.05 |
ENSRNOT00000036823
|
Ell3
|
elongation factor for RNA polymerase II 3 |
| chrX_-_104814145 | 0.05 |
ENSRNOT00000004909
|
Sytl4
|
synaptotagmin-like 4 |
| chr2_-_119537837 | 0.05 |
ENSRNOT00000015200
|
Pex5l
|
peroxisomal biogenesis factor 5-like |
| chrX_-_23649466 | 0.04 |
ENSRNOT00000059431
|
Shroom2
|
shroom family member 2 |
| chr12_+_22153983 | 0.04 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
| chr4_+_158243086 | 0.04 |
ENSRNOT00000032112
|
Ano2
|
anoctamin 2 |
| chr17_+_57075218 | 0.04 |
ENSRNOT00000089536
|
Crem
|
cAMP responsive element modulator |
| chr1_-_82120902 | 0.04 |
ENSRNOT00000027684
|
Erf
|
Ets2 repressor factor |
| chr19_-_25261911 | 0.04 |
ENSRNOT00000045180
ENSRNOT00000009806 |
Cc2d1a
|
coiled-coil and C2 domain containing 1A |
| chr8_-_22874637 | 0.04 |
ENSRNOT00000064551
ENSRNOT00000090424 |
Dock6
|
dedicator of cytokinesis 6 |
| chr1_+_221704493 | 0.04 |
ENSRNOT00000028592
|
Men1
|
menin 1 |
| chr7_+_94130852 | 0.04 |
ENSRNOT00000011485
|
Mal2
|
mal, T-cell differentiation protein 2 |
| chr14_-_86164341 | 0.04 |
ENSRNOT00000086343
ENSRNOT00000080147 |
Gck
|
glucokinase |
| chr1_-_167911961 | 0.04 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr18_+_65285318 | 0.04 |
ENSRNOT00000020431
|
Tcf4
|
transcription factor 4 |
| chr3_+_156662114 | 0.03 |
ENSRNOT00000041390
ENSRNOT00000086260 |
Top1
|
topoisomerase (DNA) I |
| chr18_-_79258570 | 0.03 |
ENSRNOT00000022401
|
Galr1
|
galanin receptor 1 |
| chr1_-_189199376 | 0.03 |
ENSRNOT00000021027
|
Umod
|
uromodulin |
| chr7_-_90318221 | 0.03 |
ENSRNOT00000050774
|
Trps1
|
transcriptional repressor GATA binding 1 |
| chr9_-_19372673 | 0.03 |
ENSRNOT00000073667
ENSRNOT00000079517 |
Clic5
|
chloride intracellular channel 5 |
| chr3_-_16537433 | 0.03 |
ENSRNOT00000048523
|
AABR07051533.2
|
|
| chr13_+_103396314 | 0.03 |
ENSRNOT00000049499
|
Slc30a10
|
solute carrier family 30, member 10 |
| chr8_+_4440876 | 0.03 |
ENSRNOT00000049325
ENSRNOT00000076529 ENSRNOT00000076748 |
Pdgfd
|
platelet derived growth factor D |
| chr1_+_198528635 | 0.03 |
ENSRNOT00000022765
|
LOC308990
|
hypothetical protein LOC308990 |
| chr4_-_150829741 | 0.03 |
ENSRNOT00000051846
ENSRNOT00000052017 |
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr1_-_80311043 | 0.03 |
ENSRNOT00000068092
|
Klc3
|
kinesin light chain 3 |
| chr5_-_147375009 | 0.03 |
ENSRNOT00000009436
|
S100pbp
|
S100P binding protein |
| chrX_+_17171605 | 0.02 |
ENSRNOT00000048236
|
Nudt10
|
nudix (nucleoside diphosphate linked moiety X)-type motif 10 |
| chr12_-_9864791 | 0.02 |
ENSRNOT00000073026
|
Gtf3a
|
general transcription factor III A |
| chr10_-_15211325 | 0.02 |
ENSRNOT00000027083
|
Rhot2
|
ras homolog family member T2 |
| chr10_+_55555089 | 0.02 |
ENSRNOT00000006597
|
Slc25a35
|
solute carrier family 25, member 35 |
| chr8_+_62341613 | 0.02 |
ENSRNOT00000066923
|
Scamp2
|
secretory carrier membrane protein 2 |
| chr4_+_65637407 | 0.02 |
ENSRNOT00000047954
|
Trim24
|
tripartite motif-containing 24 |
| chr2_-_104958034 | 0.02 |
ENSRNOT00000080699
|
Gyg1
|
glycogenin 1 |
| chr4_-_132740938 | 0.02 |
ENSRNOT00000007185
|
Rybp
|
RING1 and YY1 binding protein |
| chr5_+_13379772 | 0.02 |
ENSRNOT00000010032
|
Npbwr1
|
neuropeptides B/W receptor 1 |
| chr2_-_142885604 | 0.02 |
ENSRNOT00000031487
|
Frem2
|
Fras1 related extracellular matrix protein 2 |
| chr1_-_253185533 | 0.02 |
ENSRNOT00000067822
|
Pank1
|
pantothenate kinase 1 |
| chr2_-_188412329 | 0.02 |
ENSRNOT00000033917
ENSRNOT00000080185 |
Fdps
|
farnesyl diphosphate synthase |
| chr19_+_52077501 | 0.02 |
ENSRNOT00000079240
|
Osgin1
|
oxidative stress induced growth inhibitor 1 |
| chr13_-_42263024 | 0.02 |
ENSRNOT00000004741
|
Lypd1
|
Ly6/Plaur domain containing 1 |
| chr6_-_127630734 | 0.01 |
ENSRNOT00000089931
|
Serpina1
|
serpin family A member 1 |
| chr10_-_11901765 | 0.01 |
ENSRNOT00000009936
ENSRNOT00000075238 |
Naa60
|
N(alpha)-acetyltransferase 60, NatF catalytic subunit |
| chr1_-_205842428 | 0.01 |
ENSRNOT00000024655
|
Dhx32
|
DEAH-box helicase 32 (putative) |
| chrX_+_16337102 | 0.01 |
ENSRNOT00000003954
|
Ccnb3
|
cyclin B3 |
| chr2_-_29121104 | 0.01 |
ENSRNOT00000020543
|
Tnpo1
|
transportin 1 |
| chr10_+_91710495 | 0.01 |
ENSRNOT00000033276
|
Rprml
|
reprimo-like |
| chr9_-_85243001 | 0.01 |
ENSRNOT00000020219
|
Scg2
|
secretogranin II |
| chr1_-_79690434 | 0.01 |
ENSRNOT00000057986
ENSRNOT00000057988 |
LOC102557319
|
carcinoembryonic antigen-related cell adhesion molecule 3-like |
| chr2_+_227455722 | 0.01 |
ENSRNOT00000064809
|
Sec24d
|
SEC24 homolog D, COPII coat complex component |
| chr4_-_150829913 | 0.01 |
ENSRNOT00000041571
|
Cacna1c
|
calcium voltage-gated channel subunit alpha1 C |
| chr1_+_166125474 | 0.01 |
ENSRNOT00000091822
|
Fchsd2
|
FCH and double SH3 domains 2 |
| chr18_+_39172028 | 0.01 |
ENSRNOT00000086651
|
Kcnn2
|
potassium calcium-activated channel subfamily N member 2 |
| chrX_+_53360839 | 0.01 |
ENSRNOT00000091467
ENSRNOT00000034372 ENSRNOT00000081061 |
Dmd
|
dystrophin |
| chr10_+_11912543 | 0.01 |
ENSRNOT00000045192
|
Zfp597
|
zinc finger protein 597 |
| chr10_+_74886985 | 0.01 |
ENSRNOT00000009943
|
Mtmr4
|
myotubularin related protein 4 |
| chr9_+_71398710 | 0.01 |
ENSRNOT00000049411
|
Ccnyl1
|
cyclin Y-like 1 |
| chr7_+_83373022 | 0.01 |
ENSRNOT00000057146
ENSRNOT00000005958 |
Pkhd1l1
|
polycystic kidney and hepatic disease 1-like 1 |
| chr13_+_50873605 | 0.01 |
ENSRNOT00000004382
|
Fmod
|
fibromodulin |
| chr11_-_67037115 | 0.01 |
ENSRNOT00000003137
|
Ildr1
|
immunoglobulin-like domain containing receptor 1 |
| chr14_-_78308933 | 0.01 |
ENSRNOT00000065334
|
Crmp1
|
collapsin response mediator protein 1 |
| chr11_+_46793777 | 0.00 |
ENSRNOT00000002232
|
Adgrg7
|
adhesion G protein-coupled receptor G7 |
| chr20_+_31102476 | 0.00 |
ENSRNOT00000078719
|
Lrrc20
|
leucine rich repeat containing 20 |
| chr3_-_160139947 | 0.00 |
ENSRNOT00000014151
|
Ada
|
adenosine deaminase |
| chr1_+_193335645 | 0.00 |
ENSRNOT00000019640
|
Lcmt1
|
leucine carboxyl methyltransferase 1 |
| chr2_-_227160379 | 0.00 |
ENSRNOT00000066581
|
Usp53
|
ubiquitin specific peptidase 53 |
| chr5_+_28480023 | 0.00 |
ENSRNOT00000075067
ENSRNOT00000093754 |
LOC100912373
|
uncharacterized LOC100912373 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Scrt2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002731 | negative regulation of dendritic cell cytokine production(GO:0002731) regulation of intracellular transport of viral material(GO:1901252) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0071698 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:1903401 | L-lysine transmembrane transport(GO:1903401) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.0 | 0.0 | GO:0009732 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.0 | 0.0 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:1903763 | gap junction channel activity involved in cell communication by electrical coupling(GO:1903763) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.0 | GO:0000403 | Y-form DNA binding(GO:0000403) |