Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
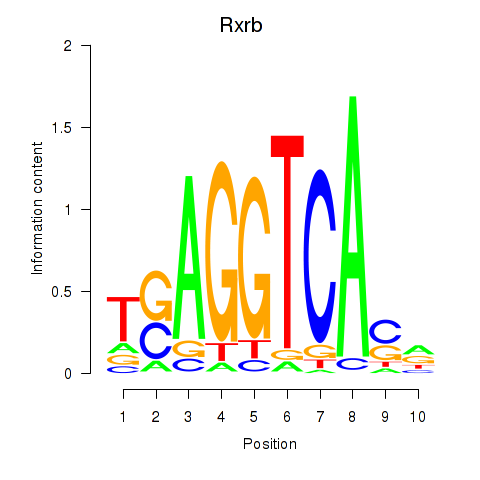
Results for Rxrb
Z-value: 0.33
Transcription factors associated with Rxrb
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxrb
|
ENSRNOG00000000464 | retinoid X receptor beta |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrb | rn6_v1_chr20_+_3827086_3827086 | -0.40 | 5.1e-01 | Click! |
Activity profile of Rxrb motif
Sorted Z-values of Rxrb motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_45215974 | 0.21 |
ENSRNOT00000010792
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr5_+_168078748 | 0.17 |
ENSRNOT00000024798
|
Uts2
|
urotensin 2 |
| chr5_+_154522119 | 0.13 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr9_+_92435896 | 0.11 |
ENSRNOT00000022901
|
Fbxo36
|
F-box protein 36 |
| chr17_+_76002275 | 0.08 |
ENSRNOT00000092665
ENSRNOT00000086701 |
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr15_+_33121273 | 0.07 |
ENSRNOT00000016020
|
Rem2
|
RRAD and GEM like GTPase 2 |
| chr6_-_136279399 | 0.06 |
ENSRNOT00000015333
|
Bag5l1
|
BCL2-associated athanogene 5-like 1 |
| chr3_+_150052483 | 0.06 |
ENSRNOT00000022265
|
RGD1561517
|
similar to chromosome 20 open reading frame 144 |
| chr5_-_152473868 | 0.05 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr6_-_136185651 | 0.05 |
ENSRNOT00000085594
|
Bag5
|
BCL2-associated athanogene 5 |
| chr12_+_24761210 | 0.05 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr6_+_136279496 | 0.05 |
ENSRNOT00000091946
|
Apopt1
|
apoptogenic 1, mitochondrial |
| chr5_-_22799090 | 0.04 |
ENSRNOT00000076426
ENSRNOT00000077044 |
Asph
|
aspartate-beta-hydroxylase |
| chr6_+_136185487 | 0.04 |
ENSRNOT00000015373
|
Apopt1
|
apoptogenic 1, mitochondrial |
| chr11_+_64601029 | 0.04 |
ENSRNOT00000004138
|
Arhgap31
|
Rho GTPase activating protein 31 |
| chr12_+_38368693 | 0.04 |
ENSRNOT00000083261
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr1_+_72635267 | 0.04 |
ENSRNOT00000090636
|
Il11
|
interleukin 11 |
| chr3_+_95715193 | 0.04 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr3_-_2434257 | 0.04 |
ENSRNOT00000013278
|
Stpg3
|
sperm-tail PG-rich repeat containing 3 |
| chr12_-_6630274 | 0.04 |
ENSRNOT00000087840
|
AABR07035194.1
|
|
| chr4_+_57715946 | 0.03 |
ENSRNOT00000079059
ENSRNOT00000034429 |
Klhdc10
|
kelch domain containing 10 |
| chr12_+_24978483 | 0.03 |
ENSRNOT00000040069
|
Eln
|
elastin |
| chr10_-_82209459 | 0.03 |
ENSRNOT00000004377
|
Spata20
|
spermatogenesis associated 20 |
| chr7_+_133400485 | 0.03 |
ENSRNOT00000006219
|
Cntn1
|
contactin 1 |
| chr15_+_34270648 | 0.03 |
ENSRNOT00000026333
|
Rnf31
|
ring finger protein 31 |
| chr2_+_198312428 | 0.03 |
ENSRNOT00000028758
|
Sf3b4
|
splicing factor 3b, subunit 4 |
| chr5_+_169357517 | 0.03 |
ENSRNOT00000000079
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr3_-_60813869 | 0.03 |
ENSRNOT00000058234
|
Atp5g3
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) |
| chr3_-_122006803 | 0.03 |
ENSRNOT00000089660
|
RGD1566226
|
similar to hypothetical protein F830045P16 |
| chrX_-_135342996 | 0.02 |
ENSRNOT00000084848
ENSRNOT00000008503 |
Aifm1
|
apoptosis inducing factor, mitochondria associated 1 |
| chr3_+_113415774 | 0.02 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr19_-_42180981 | 0.02 |
ENSRNOT00000019577
|
Pmfbp1
|
polyamine modulated factor 1 binding protein 1 |
| chr7_+_117326279 | 0.02 |
ENSRNOT00000050522
|
Spatc1
|
spermatogenesis and centriole associated 1 |
| chr3_-_149356160 | 0.02 |
ENSRNOT00000037589
ENSRNOT00000081775 |
Sun5
|
Sad1 and UNC84 domain containing 5 |
| chr6_-_122721496 | 0.02 |
ENSRNOT00000079697
|
Ptpn21
|
protein tyrosine phosphatase, non-receptor type 21 |
| chr5_-_75116490 | 0.02 |
ENSRNOT00000042788
|
Txndc8
|
thioredoxin domain containing 8 |
| chrX_-_77956352 | 0.02 |
ENSRNOT00000003296
|
Zcchc5
|
zinc finger CCHC-type containing 5 |
| chr17_+_57040023 | 0.02 |
ENSRNOT00000020204
|
Crem
|
cAMP responsive element modulator |
| chr2_+_118910587 | 0.02 |
ENSRNOT00000065518
|
Zfp639
|
zinc finger protein 639 |
| chr9_-_44786041 | 0.02 |
ENSRNOT00000067197
|
Rev1
|
REV1, DNA directed polymerase |
| chr20_+_2194709 | 0.02 |
ENSRNOT00000001017
|
Trim15
|
tripartite motif containing 15 |
| chr1_-_59347472 | 0.02 |
ENSRNOT00000017718
|
Lnpep
|
leucyl and cystinyl aminopeptidase |
| chr14_+_36047144 | 0.01 |
ENSRNOT00000003088
|
Lnx1
|
ligand of numb-protein X 1 |
| chr7_+_77066955 | 0.01 |
ENSRNOT00000008700
|
Odf1
|
outer dense fiber of sperm tails 1 |
| chr11_-_4332255 | 0.01 |
ENSRNOT00000087133
|
Cadm2
|
cell adhesion molecule 2 |
| chr14_-_108284619 | 0.01 |
ENSRNOT00000078814
|
LOC690352
|
hypothetical protein LOC690352 |
| chr4_+_113948514 | 0.01 |
ENSRNOT00000011521
|
LOC103692171
|
mannosyl-oligosaccharide glucosidase |
| chrX_-_37705263 | 0.01 |
ENSRNOT00000043666
|
Map3k15
|
mitogen-activated protein kinase kinase kinase 15 |
| chr7_-_123586919 | 0.01 |
ENSRNOT00000011484
|
Ndufa6
|
NADH:ubiquinone oxidoreductase subunit A6 |
| chr13_-_74740458 | 0.01 |
ENSRNOT00000006548
|
Tex35
|
testis expressed 35 |
| chr13_+_48679774 | 0.01 |
ENSRNOT00000075674
|
Nucks1
|
nuclear casein kinase and cyclin-dependent kinase substrate 1 |
| chr1_+_228142778 | 0.01 |
ENSRNOT00000028517
|
Mrpl16
|
mitochondrial ribosomal protein L16 |
| chr1_+_97712177 | 0.01 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chr9_+_10004805 | 0.01 |
ENSRNOT00000074117
|
Slc25a41
|
solute carrier family 25, member 41 |
| chr12_+_22142520 | 0.01 |
ENSRNOT00000038397
|
Fbxo24
|
F-box protein 24 |
| chr3_+_172856733 | 0.01 |
ENSRNOT00000009826
|
Edn3
|
endothelin 3 |
| chr4_-_120390164 | 0.01 |
ENSRNOT00000017323
ENSRNOT00000063950 |
Eefsec
|
eukaryotic elongation factor, selenocysteine-tRNA-specific |
| chr10_-_82963919 | 0.01 |
ENSRNOT00000005807
|
Dlx4
|
distal-less homeobox 4 |
| chr19_-_20508711 | 0.01 |
ENSRNOT00000032408
|
LOC680913
|
hypothetical protein LOC680913 |
| chr5_+_147069616 | 0.01 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr10_-_87535438 | 0.01 |
ENSRNOT00000086873
|
Krtap2-4
|
keratin associated protein 2-4 |
| chr8_-_55087832 | 0.00 |
ENSRNOT00000032152
|
Dlat
|
dihydrolipoamide S-acetyltransferase |
| chr13_+_93751759 | 0.00 |
ENSRNOT00000065706
|
Wdr64
|
WD repeat domain 64 |
| chr4_-_157486844 | 0.00 |
ENSRNOT00000038281
ENSRNOT00000022874 |
Cops7a
|
COP9 signalosome subunit 7A |
| chr7_-_70480724 | 0.00 |
ENSRNOT00000090057
|
Dtx3
|
deltex E3 ubiquitin ligase 3 |
| chr5_-_147303346 | 0.00 |
ENSRNOT00000009153
|
Hpca
|
hippocalcin |
| chr15_+_2740784 | 0.00 |
ENSRNOT00000067781
|
Dusp13
|
dual specificity phosphatase 13 |
| chr4_-_6062641 | 0.00 |
ENSRNOT00000074846
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr4_+_147274107 | 0.00 |
ENSRNOT00000082969
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr4_-_45332420 | 0.00 |
ENSRNOT00000083039
|
Wnt2
|
wingless-type MMTV integration site family member 2 |
| chr17_+_90802393 | 0.00 |
ENSRNOT00000003535
|
Edaradd
|
EDAR-associated death domain |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxrb
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0010841 | positive regulation of circadian sleep/wake cycle, wakefulness(GO:0010841) |
| 0.0 | 0.2 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.1 | GO:0061084 | negative regulation of protein refolding(GO:0061084) |
| 0.0 | 0.1 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.0 | GO:0021918 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.0 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |