Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Rxra
Z-value: 0.40
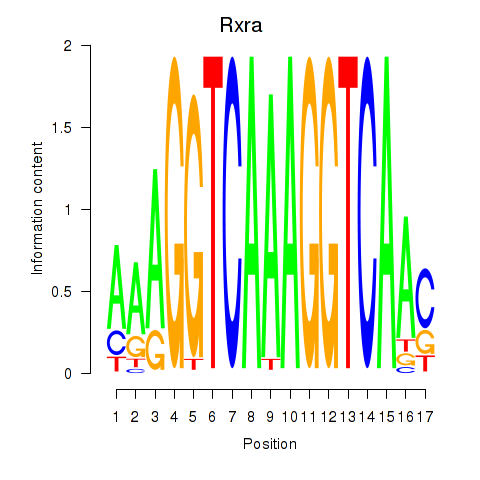
Transcription factors associated with Rxra
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rxra
|
ENSRNOG00000009446 | retinoid X receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxra | rn6_v1_chr3_+_6211789_6211789 | -0.13 | 8.4e-01 | Click! |
Activity profile of Rxra motif
Sorted Z-values of Rxra motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_91054974 | 0.20 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr3_+_124896618 | 0.18 |
ENSRNOT00000028888
|
Cds2
|
CDP-diacylglycerol synthase 2 |
| chr4_+_88832178 | 0.17 |
ENSRNOT00000088983
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr2_+_149899836 | 0.15 |
ENSRNOT00000086481
|
RGD1560324
|
similar to hypothetical protein C130079G13 |
| chr2_-_140618405 | 0.15 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr7_-_82687130 | 0.15 |
ENSRNOT00000006791
|
Tmem74
|
transmembrane protein 74 |
| chr10_+_55169282 | 0.14 |
ENSRNOT00000005423
|
Ccdc42
|
coiled-coil domain containing 42 |
| chr16_-_32868680 | 0.12 |
ENSRNOT00000015974
ENSRNOT00000082392 |
Aadat
|
aminoadipate aminotransferase |
| chr18_+_45023932 | 0.11 |
ENSRNOT00000039379
|
Fam170a
|
family with sequence similarity 170, member A |
| chr3_-_111037425 | 0.11 |
ENSRNOT00000085628
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr8_-_55696601 | 0.11 |
ENSRNOT00000016127
|
RGD1562914
|
RGD1562914 |
| chr17_+_88095830 | 0.10 |
ENSRNOT00000045984
|
Thnsl1
|
threonine synthase-like 1 |
| chr15_+_61069581 | 0.10 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr6_-_76535517 | 0.09 |
ENSRNOT00000083857
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr18_+_44468784 | 0.09 |
ENSRNOT00000031812
|
Dmxl1
|
Dmx-like 1 |
| chr14_-_92495894 | 0.09 |
ENSRNOT00000064483
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr15_-_9086282 | 0.09 |
ENSRNOT00000008989
|
Thrb
|
thyroid hormone receptor beta |
| chr1_-_239057732 | 0.09 |
ENSRNOT00000024775
|
Gda
|
guanine deaminase |
| chr2_-_47281421 | 0.09 |
ENSRNOT00000086114
|
Itga1
|
integrin subunit alpha 1 |
| chr7_-_2961873 | 0.09 |
ENSRNOT00000067441
|
Rps15-ps2
|
ribosomal protein S15, pseudogene 2 |
| chrX_+_14019961 | 0.09 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr1_+_226573105 | 0.08 |
ENSRNOT00000028074
|
Cpsf7
|
cleavage and polyadenylation specific factor 7 |
| chr12_+_6403940 | 0.08 |
ENSRNOT00000083484
|
B3glct
|
beta 3-glucosyltransferase |
| chr9_+_45901741 | 0.08 |
ENSRNOT00000059571
|
Npas2
|
neuronal PAS domain protein 2 |
| chr17_-_88095729 | 0.08 |
ENSRNOT00000025140
|
Enkur
|
enkurin, TRPC channel interacting protein |
| chr4_-_40136061 | 0.08 |
ENSRNOT00000009752
|
Bmt2
|
base methyltransferase of 25S rRNA 2 homolog |
| chr4_+_31229913 | 0.08 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr9_-_92530938 | 0.07 |
ENSRNOT00000064875
|
Slc16a14
|
solute carrier family 16, member 14 |
| chr17_+_23661429 | 0.07 |
ENSRNOT00000046523
|
Phactr1
|
phosphatase and actin regulator 1 |
| chr18_+_68983545 | 0.07 |
ENSRNOT00000085317
|
Stard6
|
StAR-related lipid transfer domain containing 6 |
| chrX_-_128268285 | 0.07 |
ENSRNOT00000009755
ENSRNOT00000081880 |
Thoc2
|
THO complex 2 |
| chr11_-_62451149 | 0.07 |
ENSRNOT00000093686
ENSRNOT00000081443 |
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_+_86914989 | 0.07 |
ENSRNOT00000085164
ENSRNOT00000081966 |
LOC100360380
|
zinc finger protein 457-like |
| chr8_+_115151627 | 0.07 |
ENSRNOT00000064252
|
Abhd14b
|
abhydrolase domain containing 14b |
| chr3_-_111037166 | 0.07 |
ENSRNOT00000017070
|
Ppp1r14d
|
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr17_+_69634890 | 0.07 |
ENSRNOT00000029049
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr12_+_19680712 | 0.07 |
ENSRNOT00000081310
|
AABR07035561.2
|
|
| chr12_-_24537313 | 0.07 |
ENSRNOT00000001975
|
Baz1b
|
bromodomain adjacent to zinc finger domain, 1B |
| chr14_+_11988671 | 0.06 |
ENSRNOT00000003207
|
Rasgef1b
|
RasGEF domain family, member 1B |
| chr6_+_78567970 | 0.06 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr3_+_100788806 | 0.06 |
ENSRNOT00000083289
ENSRNOT00000090445 |
Bdnf
|
brain-derived neurotrophic factor |
| chr2_-_102919567 | 0.06 |
ENSRNOT00000072162
|
LOC100361143
|
ribosomal protein L30-like |
| chr11_+_84396033 | 0.06 |
ENSRNOT00000002316
|
Abcc5
|
ATP binding cassette subfamily C member 5 |
| chr3_+_69549673 | 0.06 |
ENSRNOT00000043974
|
Zfp804a
|
zinc finger protein 804A |
| chr9_-_83253458 | 0.06 |
ENSRNOT00000041689
|
Epha4
|
Eph receptor A4 |
| chr13_-_80703615 | 0.06 |
ENSRNOT00000004599
|
Fmo4
|
flavin containing monooxygenase 4 |
| chr5_-_137238354 | 0.05 |
ENSRNOT00000039235
|
Szt2
|
seizure threshold 2 homolog (mouse) |
| chr7_-_117605141 | 0.05 |
ENSRNOT00000035199
|
Fbxl6
|
F-box and leucine-rich repeat protein 6 |
| chr2_-_226779440 | 0.05 |
ENSRNOT00000055669
|
Fnbp1l
|
formin binding protein 1-like |
| chr2_+_189169877 | 0.05 |
ENSRNOT00000028225
|
She
|
Src homology 2 domain containing E |
| chrX_+_74304292 | 0.05 |
ENSRNOT00000057623
|
LOC680227
|
LRRGT00193 |
| chr10_+_34975708 | 0.05 |
ENSRNOT00000064984
|
Nhp2
|
NHP2 ribonucleoprotein |
| chr1_+_47942800 | 0.05 |
ENSRNOT00000079312
|
Wtap
|
Wilms tumor 1 associated protein |
| chr17_-_56068125 | 0.05 |
ENSRNOT00000022462
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr11_+_27208564 | 0.05 |
ENSRNOT00000002158
|
Map3k7cl
|
MAP3K7 C-terminal like |
| chr2_-_178389608 | 0.05 |
ENSRNOT00000013262
|
Etfdh
|
electron transfer flavoprotein dehydrogenase |
| chr8_+_123126667 | 0.05 |
ENSRNOT00000015643
|
Osbpl10
|
oxysterol binding protein-like 10 |
| chr17_+_76306585 | 0.05 |
ENSRNOT00000065978
|
Dhtkd1
|
dehydrogenase E1 and transketolase domain containing 1 |
| chr5_+_169274785 | 0.04 |
ENSRNOT00000082586
|
Plekhg5
|
pleckstrin homology and RhoGEF domain containing G5 |
| chr12_-_6078411 | 0.04 |
ENSRNOT00000001197
|
Rxfp2
|
relaxin/insulin-like family peptide receptor 2 |
| chr15_+_39945095 | 0.04 |
ENSRNOT00000016826
|
Shisa2
|
shisa family member 2 |
| chr6_+_59757397 | 0.04 |
ENSRNOT00000047001
|
AABR07064000.1
|
|
| chr10_+_3227160 | 0.04 |
ENSRNOT00000088075
|
Ntan1
|
N-terminal asparagine amidase |
| chr4_+_2340148 | 0.04 |
ENSRNOT00000072890
|
Nom1
|
nucleolar protein with MIF4G domain 1 |
| chr4_+_183896303 | 0.04 |
ENSRNOT00000055437
|
RGD1309621
|
similar to hypothetical protein FLJ10652 |
| chr3_+_161272385 | 0.04 |
ENSRNOT00000021052
|
Zswim3
|
zinc finger, SWIM-type containing 3 |
| chr5_+_144581427 | 0.04 |
ENSRNOT00000015227
|
Clspn
|
claspin |
| chr8_-_48597867 | 0.04 |
ENSRNOT00000077958
|
Nlrx1
|
NLR family member X1 |
| chr1_+_15620653 | 0.04 |
ENSRNOT00000017401
|
Map7
|
microtubule-associated protein 7 |
| chr20_+_3827086 | 0.04 |
ENSRNOT00000081588
|
Rxrb
|
retinoid X receptor beta |
| chr9_+_100932932 | 0.04 |
ENSRNOT00000065058
|
Ing5
|
inhibitor of growth family, member 5 |
| chr9_-_13311924 | 0.04 |
ENSRNOT00000015584
|
Kif6
|
kinesin family member 6 |
| chr10_-_18443934 | 0.04 |
ENSRNOT00000059895
ENSRNOT00000080021 |
Ranbp17
|
RAN binding protein 17 |
| chr4_+_34282625 | 0.04 |
ENSRNOT00000011138
ENSRNOT00000086735 |
Glcci1
|
glucocorticoid induced 1 |
| chr1_+_15834779 | 0.04 |
ENSRNOT00000079069
ENSRNOT00000083012 |
Bclaf1
|
BCL2-associated transcription factor 1 |
| chr1_+_257076221 | 0.04 |
ENSRNOT00000044895
|
Slc35g1
|
solute carrier family 35, member G1 |
| chr9_+_81783349 | 0.04 |
ENSRNOT00000021548
|
Cnot9
|
CCR4-NOT transcription complex subunit 9 |
| chr3_-_52447622 | 0.04 |
ENSRNOT00000083552
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr2_+_196304492 | 0.04 |
ENSRNOT00000028640
|
Lysmd1
|
LysM domain containing 1 |
| chr1_+_187149453 | 0.04 |
ENSRNOT00000082738
|
Xylt1
|
xylosyltransferase 1 |
| chr15_+_675085 | 0.04 |
ENSRNOT00000091318
|
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr1_+_61733805 | 0.04 |
ENSRNOT00000086029
|
LOC682206
|
similar to Zinc finger protein 208 |
| chr18_+_59096643 | 0.04 |
ENSRNOT00000025325
|
Wdr7
|
WD repeat domain 7 |
| chrX_+_20351486 | 0.04 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr3_-_45210474 | 0.04 |
ENSRNOT00000091777
|
Ccdc148
|
coiled-coil domain containing 148 |
| chr2_-_199971965 | 0.04 |
ENSRNOT00000087026
|
Pde4dip
|
phosphodiesterase 4D interacting protein |
| chr5_-_171710312 | 0.04 |
ENSRNOT00000076044
ENSRNOT00000071280 ENSRNOT00000072059 ENSRNOT00000077015 |
Prdm16
|
PR/SET domain 16 |
| chr8_-_14880644 | 0.04 |
ENSRNOT00000015977
|
Fat3
|
FAT atypical cadherin 3 |
| chr16_+_74810938 | 0.04 |
ENSRNOT00000058091
|
Nek5
|
NIMA-related kinase 5 |
| chr2_-_89498395 | 0.04 |
ENSRNOT00000068507
|
AABR07009224.1
|
|
| chr10_-_14299167 | 0.03 |
ENSRNOT00000042066
|
Mapk8ip3
|
mitogen-activated protein kinase 8 interacting protein 3 |
| chr1_-_31122093 | 0.03 |
ENSRNOT00000016712
|
NEWGENE_1307525
|
SOGA family member 3 |
| chr2_+_166403265 | 0.03 |
ENSRNOT00000012417
|
Nmd3
|
NMD3 ribosome export adaptor |
| chr1_+_198655742 | 0.03 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr14_+_1462358 | 0.03 |
ENSRNOT00000077243
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr1_-_71173216 | 0.03 |
ENSRNOT00000020523
|
Zfp28
|
zinc finger protein 28 |
| chr16_-_22853022 | 0.03 |
ENSRNOT00000050775
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr6_+_44230985 | 0.03 |
ENSRNOT00000005858
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr1_-_37932084 | 0.03 |
ENSRNOT00000022527
|
LOC102547287
|
zinc finger protein 728-like |
| chr8_+_99894762 | 0.03 |
ENSRNOT00000081768
|
Plscr4
|
phospholipid scramblase 4 |
| chr6_+_123034304 | 0.03 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
| chr11_+_47146308 | 0.03 |
ENSRNOT00000002191
|
Cep97
|
centrosomal protein 97 |
| chr9_+_92681078 | 0.03 |
ENSRNOT00000034152
|
Sp100
|
SP100 nuclear antigen |
| chr2_+_166402838 | 0.03 |
ENSRNOT00000064244
|
Nmd3
|
NMD3 ribosome export adaptor |
| chr2_-_111793326 | 0.03 |
ENSRNOT00000092660
|
Nlgn1
|
neuroligin 1 |
| chr3_+_176494768 | 0.03 |
ENSRNOT00000040270
|
Col20a1
|
collagen type XX alpha 1 chain |
| chrX_+_29562165 | 0.03 |
ENSRNOT00000006074
|
Ofd1
|
OFD1, centriole and centriolar satellite protein |
| chr1_-_100231460 | 0.03 |
ENSRNOT00000032200
|
Acpt
|
acid phosphatase, testicular |
| chr6_+_26537707 | 0.03 |
ENSRNOT00000050102
|
Zfp513
|
zinc finger protein 513 |
| chrX_-_77295426 | 0.03 |
ENSRNOT00000090833
|
Taf9b
|
TATA-box binding protein associated factor 9b |
| chr8_+_40009691 | 0.03 |
ENSRNOT00000042679
|
Vsig2
|
V-set and immunoglobulin domain containing 2 |
| chr10_-_67285617 | 0.03 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chr8_-_48674748 | 0.03 |
ENSRNOT00000014127
|
Hmbs
|
hydroxymethylbilane synthase |
| chr19_+_22450030 | 0.03 |
ENSRNOT00000021739
|
Neto2
|
neuropilin and tolloid like 2 |
| chr14_-_83603003 | 0.03 |
ENSRNOT00000050005
ENSRNOT00000057673 |
Limk2
|
LIM domain kinase 2 |
| chr10_-_102423084 | 0.03 |
ENSRNOT00000065145
|
Sdk2
|
sidekick cell adhesion molecule 2 |
| chr12_-_31629881 | 0.03 |
ENSRNOT00000001238
|
Piwil1
|
piwi-like RNA-mediated gene silencing 1 |
| chr20_+_18833481 | 0.03 |
ENSRNOT00000080846
|
Bicc1
|
BicC family RNA binding protein 1 |
| chr1_+_221236773 | 0.03 |
ENSRNOT00000051979
|
Slc25a45
|
solute carrier family 25, member 45 |
| chr4_+_114854458 | 0.03 |
ENSRNOT00000013312
|
LOC500227
|
hypothetical gene supported by BC079424 |
| chr14_-_78377825 | 0.03 |
ENSRNOT00000068104
|
AABR07015812.1
|
|
| chr1_+_85192174 | 0.02 |
ENSRNOT00000026966
|
Pak4
|
p21 (RAC1) activated kinase 4 |
| chr10_+_109852036 | 0.02 |
ENSRNOT00000054944
|
Aspscr1
|
ASPSCR1, UBX domain containing tether for SLC2A4 |
| chr17_+_55065621 | 0.02 |
ENSRNOT00000086342
|
Zfp438
|
zinc finger protein 438 |
| chr6_-_26792808 | 0.02 |
ENSRNOT00000010403
|
Abhd1
|
abhydrolase domain containing 1 |
| chr11_-_83882117 | 0.02 |
ENSRNOT00000073735
|
LOC100912534
|
DNA-directed RNA polymerases I, II, and III subunit RPABC3-like |
| chr1_+_213676954 | 0.02 |
ENSRNOT00000050551
|
Nlrp6
|
NLR family, pyrin domain containing 6 |
| chr6_+_106496992 | 0.02 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr4_+_117215064 | 0.02 |
ENSRNOT00000020909
|
Smyd5
|
SMYD family member 5 |
| chr1_-_140835151 | 0.02 |
ENSRNOT00000032174
|
Hapln3
|
hyaluronan and proteoglycan link protein 3 |
| chr7_+_11356118 | 0.02 |
ENSRNOT00000041325
|
Atcay
|
ATCAY, caytaxin |
| chr1_+_86973745 | 0.02 |
ENSRNOT00000078156
|
Rinl
|
Ras and Rab interactor-like |
| chr7_+_140788987 | 0.02 |
ENSRNOT00000086611
|
Kcnh3
|
potassium voltage-gated channel subfamily H member 3 |
| chr8_+_114876146 | 0.02 |
ENSRNOT00000075966
|
Wdr82
|
WD repeat domain 82 |
| chr14_-_45376127 | 0.02 |
ENSRNOT00000059247
|
AABR07015015.2
|
|
| chr20_-_45259928 | 0.02 |
ENSRNOT00000087226
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr3_+_171213936 | 0.02 |
ENSRNOT00000031586
|
Pck1
|
phosphoenolpyruvate carboxykinase 1 |
| chr1_+_86914137 | 0.02 |
ENSRNOT00000027006
|
Fbxo17
|
F-box protein 17 |
| chr4_+_29978739 | 0.02 |
ENSRNOT00000011756
|
Ppp1r9a
|
protein phosphatase 1, regulatory subunit 9A |
| chr20_+_30915213 | 0.02 |
ENSRNOT00000000681
|
Prf1
|
perforin 1 |
| chrX_-_1704033 | 0.02 |
ENSRNOT00000051956
|
Usp11
|
ubiquitin specific peptidase 11 |
| chr9_+_79659251 | 0.02 |
ENSRNOT00000021656
|
Xrcc5
|
X-ray repair cross complementing 5 |
| chr8_+_2659865 | 0.02 |
ENSRNOT00000088553
ENSRNOT00000010243 |
Casp12
|
caspase 12 |
| chrX_+_45637415 | 0.02 |
ENSRNOT00000050544
|
RGD1563378
|
similar to ferritin heavy polypeptide-like 17 |
| chr2_+_27365148 | 0.02 |
ENSRNOT00000021549
|
Col4a3bp
|
collagen type IV alpha 3 binding protein |
| chr16_-_19349080 | 0.02 |
ENSRNOT00000038494
|
Hsh2d
|
hematopoietic SH2 domain containing |
| chr5_-_2803855 | 0.02 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr13_+_83721300 | 0.02 |
ENSRNOT00000082677
|
Adcy10
|
adenylate cyclase 10 (soluble) |
| chr18_-_11858744 | 0.02 |
ENSRNOT00000061417
ENSRNOT00000082891 |
Dsc2
|
desmocollin 2 |
| chr8_-_83693472 | 0.02 |
ENSRNOT00000015947
|
Fam83b
|
family with sequence similarity 83, member B |
| chr5_+_173660921 | 0.02 |
ENSRNOT00000066561
|
Noc2l
|
NOC2-like nucleolar associated transcriptional repressor |
| chr19_+_37330930 | 0.02 |
ENSRNOT00000022439
|
Plekhg4
|
pleckstrin homology and RhoGEF domain containing G4 |
| chr12_+_51937175 | 0.02 |
ENSRNOT00000056780
|
Pus1
|
pseudouridylate synthase 1 |
| chr1_+_219144205 | 0.02 |
ENSRNOT00000083942
|
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr3_+_141068023 | 0.02 |
ENSRNOT00000036527
|
Kiz
|
kizuna centrosomal protein |
| chr20_-_2159631 | 0.02 |
ENSRNOT00000061558
|
Trim31
|
tripartite motif-containing 31 |
| chr19_+_49637016 | 0.01 |
ENSRNOT00000016880
|
Bco1
|
beta-carotene oxygenase 1 |
| chr11_+_61531571 | 0.01 |
ENSRNOT00000093467
ENSRNOT00000002727 |
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr2_-_90568486 | 0.01 |
ENSRNOT00000059380
|
LOC100910852
|
uncharacterized LOC100910852 |
| chr10_-_88143965 | 0.01 |
ENSRNOT00000080167
|
Ka11
|
type I keratin KA11 |
| chr13_-_47440682 | 0.01 |
ENSRNOT00000037679
ENSRNOT00000005729 ENSRNOT00000050354 ENSRNOT00000050859 |
Pfkfb2
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr13_+_83681322 | 0.01 |
ENSRNOT00000004206
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr10_-_104748003 | 0.01 |
ENSRNOT00000042372
ENSRNOT00000046754 |
Acox1
|
acyl-CoA oxidase 1 |
| chr16_-_21348391 | 0.01 |
ENSRNOT00000083537
|
Lpar2
|
lysophosphatidic acid receptor 2 |
| chr7_-_136853957 | 0.01 |
ENSRNOT00000008985
|
Nell2
|
neural EGFL like 2 |
| chr3_-_10371240 | 0.01 |
ENSRNOT00000012075
|
Ass1
|
argininosuccinate synthase 1 |
| chr3_+_114129589 | 0.01 |
ENSRNOT00000056119
|
Terb2
|
telomere repeat binding bouquet formation protein 2 |
| chr1_+_80954858 | 0.01 |
ENSRNOT00000030440
|
Zfp112
|
zinc finger protein 112 |
| chr19_-_49510901 | 0.01 |
ENSRNOT00000082929
ENSRNOT00000079969 |
LOC687399
|
hypothetical protein LOC687399 |
| chr1_-_212633304 | 0.01 |
ENSRNOT00000025611
|
Olr286
|
olfactory receptor 286 |
| chr2_-_250862419 | 0.01 |
ENSRNOT00000017943
|
Clca4
|
chloride channel accessory 4 |
| chr8_+_127171537 | 0.01 |
ENSRNOT00000078625
ENSRNOT00000051290 ENSRNOT00000087753 |
Golga4
|
golgin A4 |
| chr4_+_5644260 | 0.01 |
ENSRNOT00000041876
|
Actr3b
|
ARP3 actin related protein 3 homolog B |
| chr9_-_69953182 | 0.01 |
ENSRNOT00000015852
|
Ndufs1
|
NADH dehydrogenase (ubiquinone) Fe-S protein 1 |
| chr15_+_5306459 | 0.01 |
ENSRNOT00000092079
|
LOC108348081
|
CD99 antigen-like protein 2 |
| chr10_+_96639924 | 0.01 |
ENSRNOT00000004756
|
Apoh
|
apolipoprotein H |
| chr10_-_85289777 | 0.01 |
ENSRNOT00000055427
ENSRNOT00000014863 |
Gpr179
|
G protein-coupled receptor 179 |
| chr14_-_102293677 | 0.01 |
ENSRNOT00000008135
|
Etaa1
|
Ewing tumor-associated antigen 1 |
| chr8_+_56451240 | 0.01 |
ENSRNOT00000085278
|
LOC100359687
|
mitochondrial ribosomal protein L1-like |
| chr7_+_12798868 | 0.01 |
ENSRNOT00000038553
|
Prss57
|
protease, serine, 57 |
| chr1_+_229066045 | 0.01 |
ENSRNOT00000016454
|
Glyat
|
glycine-N-acyltransferase |
| chr1_+_220243473 | 0.01 |
ENSRNOT00000084650
ENSRNOT00000027091 |
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr2_+_40554146 | 0.01 |
ENSRNOT00000015138
|
Pde4d
|
phosphodiesterase 4D |
| chr1_-_219144610 | 0.01 |
ENSRNOT00000023526
|
Ndufs8
|
NADH:ubiquinone oxidoreductase core subunit S8 |
| chr8_+_39878955 | 0.01 |
ENSRNOT00000060300
|
LOC100911068
|
roundabout homolog 4-like |
| chr3_+_56056925 | 0.01 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr8_-_119265157 | 0.01 |
ENSRNOT00000056100
|
Rtp3
|
receptor (chemosensory) transporter protein 3 |
| chr1_+_97712177 | 0.01 |
ENSRNOT00000027745
|
MGC114499
|
similar to RIKEN cDNA 4930433I11 gene |
| chr8_+_117246376 | 0.01 |
ENSRNOT00000074493
|
Ccdc71
|
coiled-coil domain containing 71 |
| chr5_-_102743417 | 0.01 |
ENSRNOT00000067389
|
Bnc2
|
basonuclin 2 |
| chr7_+_71004417 | 0.01 |
ENSRNOT00000005575
|
Myo1a
|
myosin IA |
| chr17_-_49990982 | 0.01 |
ENSRNOT00000018458
ENSRNOT00000080628 |
Mplkip
|
M-phase specific PLK1 interacting protein |
| chr6_-_122239614 | 0.01 |
ENSRNOT00000005015
|
Galc
|
galactosylceramidase |
| chr18_+_46148849 | 0.01 |
ENSRNOT00000026724
|
Prr16
|
proline rich 16 |
| chr14_+_1463359 | 0.01 |
ENSRNOT00000070834
|
Csf2ra
|
colony stimulating factor 2 receptor alpha subunit |
| chr2_+_218834034 | 0.01 |
ENSRNOT00000018367
|
Dph5
|
diphthamide biosynthesis 5 |
| chr7_-_116920507 | 0.01 |
ENSRNOT00000048363
|
Mroh6
|
maestro heat-like repeat family member 6 |
| chr10_-_56624526 | 0.01 |
ENSRNOT00000024973
|
Acadvl
|
acyl-CoA dehydrogenase, very long chain |
| chr16_-_81834945 | 0.01 |
ENSRNOT00000037806
|
F7
|
coagulation factor VII |
| chr10_+_11810926 | 0.01 |
ENSRNOT00000036205
ENSRNOT00000036189 |
Nlrc3
|
NLR family, CARD domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rxra
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1904612 | response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:2001106 | fasciculation of motor neuron axon(GO:0097156) regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.2 | GO:0006655 | phosphatidylglycerol biosynthetic process(GO:0006655) |
| 0.0 | 0.1 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0060082 | eye blink reflex(GO:0060082) |
| 0.0 | 0.0 | GO:2000688 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 0.0 | 0.1 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.0 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004605 | phosphatidate cytidylyltransferase activity(GO:0004605) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0036137 | kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) |
| 0.0 | 0.1 | GO:0016433 | rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.0 | GO:0032551 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.0 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.0 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0042731 | PH domain binding(GO:0042731) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.1 | REACTOME TRYPTOPHAN CATABOLISM | Genes involved in Tryptophan catabolism |
| 0.0 | 0.1 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |