Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Runx2_Bcl11a
Z-value: 0.90
Transcription factors associated with Runx2_Bcl11a
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
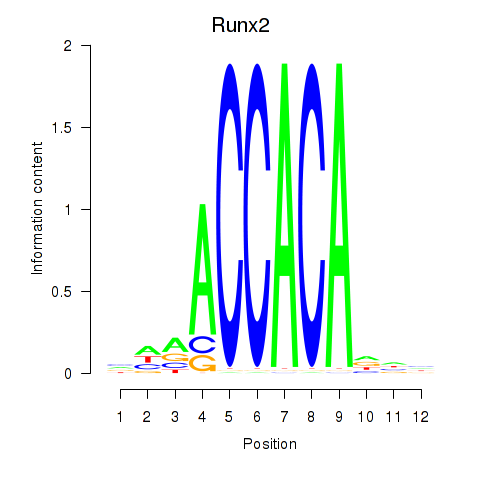
Runx2
|
ENSRNOG00000020193 | runt-related transcription factor 2 |
|
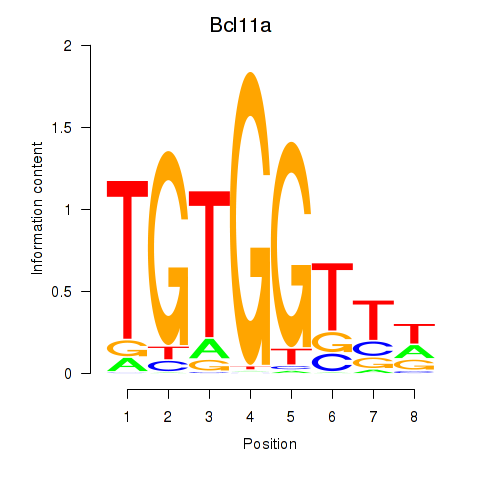
Bcl11a
|
ENSRNOG00000007049 | B-cell CLL/lymphoma 11A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx2 | rn6_v1_chr9_+_18564927_18564927 | 0.57 | 3.2e-01 | Click! |
| Bcl11a | rn6_v1_chr14_+_108826831_108826939 | 0.55 | 3.4e-01 | Click! |
Activity profile of Runx2_Bcl11a motif
Sorted Z-values of Runx2_Bcl11a motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_14348046 | 0.63 |
ENSRNOT00000018630
|
Cdhr1
|
cadherin-related family member 1 |
| chr11_-_33003021 | 0.42 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr1_+_91152635 | 0.36 |
ENSRNOT00000073438
|
LOC100912070
|
dermokine-like |
| chr6_+_143938040 | 0.33 |
ENSRNOT00000005876
|
Vipr2
|
vasoactive intestinal peptide receptor 2 |
| chr13_+_48455923 | 0.32 |
ENSRNOT00000009280
|
Rab7b
|
Rab7b, member RAS oncogene family |
| chr10_+_73868943 | 0.32 |
ENSRNOT00000081012
|
Tubd1
|
tubulin, delta 1 |
| chr2_-_187742747 | 0.31 |
ENSRNOT00000026530
|
Bglap
|
bone gamma-carboxyglutamate protein |
| chr10_-_38985466 | 0.30 |
ENSRNOT00000010121
|
Il13
|
interleukin 13 |
| chr2_+_210381829 | 0.30 |
ENSRNOT00000024611
|
Alx3
|
ALX homeobox 3 |
| chr10_+_69423086 | 0.30 |
ENSRNOT00000000256
|
Ccl7
|
C-C motif chemokine ligand 7 |
| chr1_-_82004538 | 0.29 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr7_-_143967484 | 0.28 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr1_-_178367828 | 0.27 |
ENSRNOT00000050823
|
AABR07071968.1
|
|
| chr19_-_43911057 | 0.27 |
ENSRNOT00000026017
|
Ctrb1
|
chymotrypsinogen B1 |
| chr1_+_219173002 | 0.26 |
ENSRNOT00000024018
|
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr19_-_28751584 | 0.25 |
ENSRNOT00000079270
|
AABR07043456.1
|
|
| chr20_-_22459025 | 0.24 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr18_-_37096132 | 0.24 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr8_-_133002201 | 0.24 |
ENSRNOT00000008772
|
Ccr1
|
C-C motif chemokine receptor 1 |
| chr1_+_88875375 | 0.24 |
ENSRNOT00000028284
|
Tyrobp
|
Tyro protein tyrosine kinase binding protein |
| chrX_-_138148967 | 0.23 |
ENSRNOT00000033968
|
Frmd7
|
FERM domain containing 7 |
| chr11_+_88122271 | 0.23 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr10_-_56270640 | 0.23 |
ENSRNOT00000056918
|
Cd68
|
Cd68 molecule |
| chr16_-_75309176 | 0.23 |
ENSRNOT00000018427
|
Defb1
|
defensin beta 1 |
| chr3_-_6626284 | 0.22 |
ENSRNOT00000012494
|
Fcnb
|
ficolin B |
| chr5_-_151898022 | 0.22 |
ENSRNOT00000000133
|
Pigv
|
phosphatidylinositol glycan anchor biosynthesis, class V |
| chr1_-_226887156 | 0.21 |
ENSRNOT00000054809
ENSRNOT00000028347 |
Cd6
|
Cd6 molecule |
| chr15_-_34612432 | 0.21 |
ENSRNOT00000090206
|
Mcpt1l1
|
mast cell protease 1-like 1 |
| chr5_+_8459660 | 0.20 |
ENSRNOT00000007491
|
Cpa6
|
carboxypeptidase A6 |
| chr1_-_37990007 | 0.20 |
ENSRNOT00000081054
|
AABR07001100.1
|
|
| chr2_+_195582781 | 0.20 |
ENSRNOT00000066020
|
Them5
|
thioesterase superfamily member 5 |
| chr3_-_67668772 | 0.19 |
ENSRNOT00000010247
|
Frzb
|
frizzled-related protein |
| chr10_-_83655182 | 0.19 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr11_-_29710849 | 0.19 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr9_-_15700235 | 0.19 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr1_+_44311513 | 0.19 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr1_-_169463760 | 0.18 |
ENSRNOT00000023100
|
Trim30c
|
tripartite motif-containing 30C |
| chr17_-_90315492 | 0.18 |
ENSRNOT00000070807
|
Gng4
|
G protein subunit gamma 4 |
| chr20_+_3189473 | 0.18 |
ENSRNOT00000047439
|
RT1-T24-4
|
RT1 class I, locus T24, gene 4 |
| chr16_-_75340360 | 0.18 |
ENSRNOT00000018501
|
Defa5
|
defensin alpha 5 |
| chr13_+_52662996 | 0.18 |
ENSRNOT00000047682
|
Tnnt2
|
troponin T2, cardiac type |
| chr4_-_156276304 | 0.18 |
ENSRNOT00000078725
|
Clec4e
|
C-type lectin domain family 4, member E |
| chr17_+_49417067 | 0.18 |
ENSRNOT00000090024
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr14_+_70780623 | 0.18 |
ENSRNOT00000083871
ENSRNOT00000058803 |
Ldb2
|
LIM domain binding 2 |
| chr10_-_107376645 | 0.17 |
ENSRNOT00000046213
|
Cep295nl
|
CEP295 N-terminal like |
| chr13_-_111917587 | 0.16 |
ENSRNOT00000007649
|
Traf3ip3
|
TRAF3 interacting protein 3 |
| chr5_+_25253010 | 0.16 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr9_+_18564927 | 0.16 |
ENSRNOT00000061014
|
Runx2
|
runt-related transcription factor 2 |
| chr1_+_101214593 | 0.16 |
ENSRNOT00000028086
|
Tead2
|
TEA domain transcription factor 2 |
| chr15_-_34693034 | 0.16 |
ENSRNOT00000083314
|
Mcpt8
|
mast cell protease 8 |
| chr16_+_2706428 | 0.16 |
ENSRNOT00000077117
|
Il17rd
|
interleukin 17 receptor D |
| chr14_-_18853315 | 0.16 |
ENSRNOT00000003794
|
Ppbp
|
pro-platelet basic protein |
| chr14_-_39112600 | 0.16 |
ENSRNOT00000003170
|
Gabrb1
|
gamma-aminobutyric acid type A receptor beta 1 subunit |
| chr11_+_86520992 | 0.15 |
ENSRNOT00000040954
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr1_+_81208212 | 0.15 |
ENSRNOT00000026288
|
Lypd5
|
Ly6/Plaur domain containing 5 |
| chrX_-_134719097 | 0.15 |
ENSRNOT00000068478
|
Smarca1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 |
| chr9_-_63641400 | 0.15 |
ENSRNOT00000087684
|
Satb2
|
SATB homeobox 2 |
| chr6_+_106039991 | 0.15 |
ENSRNOT00000088917
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr1_+_98398660 | 0.15 |
ENSRNOT00000047473
|
Cd33
|
CD33 molecule |
| chr4_+_66670618 | 0.15 |
ENSRNOT00000010796
|
Tbxas1
|
thromboxane A synthase 1 |
| chr1_-_261446570 | 0.15 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr5_+_25725683 | 0.14 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr3_+_67849966 | 0.14 |
ENSRNOT00000057826
|
Dusp19
|
dual specificity phosphatase 19 |
| chr1_+_219199158 | 0.14 |
ENSRNOT00000024034
|
LOC688778
|
similar to fatty aldehyde dehydrogenase-like |
| chr7_+_12798868 | 0.14 |
ENSRNOT00000038553
|
Prss57
|
protease, serine, 57 |
| chr5_-_147784311 | 0.14 |
ENSRNOT00000074172
|
Fam167b
|
family with sequence similarity 167, member B |
| chr5_-_152423686 | 0.14 |
ENSRNOT00000056143
|
Umodl
|
Uromoduilin-like |
| chr1_-_213818222 | 0.14 |
ENSRNOT00000054875
|
Ifitm6
|
interferon induced transmembrane protein 6 |
| chr2_+_86951776 | 0.14 |
ENSRNOT00000087275
|
AABR07009105.1
|
|
| chr9_+_67546408 | 0.14 |
ENSRNOT00000013701
|
Cd28
|
Cd28 molecule |
| chr8_-_117237229 | 0.14 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr8_+_2604962 | 0.13 |
ENSRNOT00000009993
|
Casp1
|
caspase 1 |
| chr13_+_75059927 | 0.13 |
ENSRNOT00000080801
|
LOC680254
|
hypothetical protein LOC680254 |
| chr1_+_65576535 | 0.13 |
ENSRNOT00000026575
|
Slc27a5
|
solute carrier family 27 member 5 |
| chr3_+_110466790 | 0.13 |
ENSRNOT00000089434
|
Pak6
|
p21 (RAC1) activated kinase 6 |
| chr1_+_170242846 | 0.13 |
ENSRNOT00000023751
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr3_+_137154086 | 0.13 |
ENSRNOT00000034252
|
Otor
|
otoraplin |
| chr10_+_75087892 | 0.13 |
ENSRNOT00000065910
|
Mpo
|
myeloperoxidase |
| chrX_+_159081445 | 0.13 |
ENSRNOT00000056688
|
AABR07042542.1
|
|
| chr1_-_170175350 | 0.13 |
ENSRNOT00000023349
|
Olr201
|
olfactory receptor 201 |
| chr2_+_55835151 | 0.13 |
ENSRNOT00000018634
|
Fyb
|
FYN binding protein |
| chr1_+_190462327 | 0.13 |
ENSRNOT00000030732
|
LOC691519
|
similar to ankyrin repeat domain 26 |
| chr2_-_104955807 | 0.12 |
ENSRNOT00000014837
|
Gyg1
|
glycogenin 1 |
| chr8_+_49621568 | 0.12 |
ENSRNOT00000021949
|
Tmprss13
|
transmembrane protease, serine 13 |
| chr4_+_96831880 | 0.12 |
ENSRNOT00000068400
|
RSA-14-44
|
RSA-14-44 protein |
| chr1_-_131460473 | 0.12 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr3_+_66673246 | 0.12 |
ENSRNOT00000081338
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_+_44096061 | 0.12 |
ENSRNOT00000018438
|
Ankrd55
|
ankyrin repeat domain 55 |
| chr1_-_169456098 | 0.12 |
ENSRNOT00000030827
|
Trim30c
|
tripartite motif-containing 30C |
| chr14_-_46054022 | 0.11 |
ENSRNOT00000002982
|
LOC498368
|
similar to RIKEN cDNA 0610040J01 |
| chr4_-_78208767 | 0.11 |
ENSRNOT00000033918
|
Rarres2
|
retinoic acid receptor responder 2 |
| chr2_-_106703401 | 0.11 |
ENSRNOT00000043933
|
AABR07009638.1
|
|
| chr9_+_74124016 | 0.11 |
ENSRNOT00000019023
|
Cps1
|
carbamoyl-phosphate synthase 1 |
| chr4_-_132171153 | 0.11 |
ENSRNOT00000015058
ENSRNOT00000015075 |
Prok2
|
prokineticin 2 |
| chr7_-_51515131 | 0.11 |
ENSRNOT00000006773
ENSRNOT00000041473 ENSRNOT00000050037 |
Ppp1r12a
|
protein phosphatase 1, regulatory subunit 12A |
| chr18_+_55505993 | 0.11 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr2_-_147392062 | 0.11 |
ENSRNOT00000021535
|
Tm4sf1
|
transmembrane 4 L six family member 1 |
| chr18_-_28438654 | 0.11 |
ENSRNOT00000036301
|
Mzb1
|
marginal zone B and B1 cell-specific protein |
| chr5_-_57632177 | 0.11 |
ENSRNOT00000080787
ENSRNOT00000092581 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr14_+_10692764 | 0.11 |
ENSRNOT00000003012
|
LOC100910270
|
uncharacterized LOC100910270 |
| chr4_+_179398621 | 0.11 |
ENSRNOT00000049474
ENSRNOT00000067506 |
Lrmp
|
lymphoid-restricted membrane protein |
| chr5_-_135994848 | 0.11 |
ENSRNOT00000067675
|
Btbd19
|
BTB domain containing 19 |
| chr11_-_81444375 | 0.11 |
ENSRNOT00000058479
ENSRNOT00000078131 ENSRNOT00000080949 ENSRNOT00000080562 ENSRNOT00000084867 |
Kng1
|
kininogen 1 |
| chr20_+_27129291 | 0.11 |
ENSRNOT00000031053
|
AABR07044933.1
|
|
| chr2_-_122949241 | 0.11 |
ENSRNOT00000039615
|
Qrfpr
|
pyroglutamylated RFamide peptide receptor |
| chr4_-_84131030 | 0.11 |
ENSRNOT00000012196
|
Cpvl
|
carboxypeptidase, vitellogenic-like |
| chr2_-_250241590 | 0.11 |
ENSRNOT00000077221
ENSRNOT00000067502 |
Lmo4
|
LIM domain only 4 |
| chr9_+_71247781 | 0.11 |
ENSRNOT00000049654
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr1_-_167700332 | 0.11 |
ENSRNOT00000092890
|
Trim21
|
tripartite motif-containing 21 |
| chr1_+_1771710 | 0.11 |
ENSRNOT00000080138
ENSRNOT00000073528 |
Nup43
|
nucleoporin 43 |
| chr17_+_49714294 | 0.11 |
ENSRNOT00000018190
|
Rala
|
RAS like proto-oncogene A |
| chr7_-_143966863 | 0.10 |
ENSRNOT00000018828
|
Sp7
|
Sp7 transcription factor |
| chr20_-_5806097 | 0.10 |
ENSRNOT00000000611
|
Clps
|
colipase |
| chr5_+_58661049 | 0.10 |
ENSRNOT00000078274
|
Unc13b
|
unc-13 homolog B |
| chr15_+_33074441 | 0.10 |
ENSRNOT00000075610
|
Mmp14
|
matrix metallopeptidase 14 |
| chr5_+_135562034 | 0.10 |
ENSRNOT00000056967
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr14_-_5378726 | 0.10 |
ENSRNOT00000002896
|
Lrrc8c
|
leucine rich repeat containing 8 family, member C |
| chr11_+_83868655 | 0.10 |
ENSRNOT00000072402
|
NEWGENE_621438
|
thrombopoietin |
| chr11_+_31694339 | 0.10 |
ENSRNOT00000002779
|
Ifngr2
|
interferon gamma receptor 2 |
| chr9_+_95161157 | 0.10 |
ENSRNOT00000071200
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr2_+_198965685 | 0.10 |
ENSRNOT00000000107
ENSRNOT00000091578 |
Pdzk1
|
PDZ domain containing 1 |
| chr3_+_66673071 | 0.10 |
ENSRNOT00000034769
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr20_+_3156170 | 0.10 |
ENSRNOT00000082880
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr14_+_13192347 | 0.10 |
ENSRNOT00000000092
|
Antxr2
|
anthrax toxin receptor 2 |
| chr7_-_145450233 | 0.10 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr1_+_212558257 | 0.10 |
ENSRNOT00000024912
|
Prap1
|
proline-rich acidic protein 1 |
| chr20_-_5805627 | 0.10 |
ENSRNOT00000085996
|
Clps
|
colipase |
| chr8_-_3272306 | 0.10 |
ENSRNOT00000040365
|
AABR07068955.1
|
|
| chr8_-_116531784 | 0.10 |
ENSRNOT00000024529
|
Rbm5
|
RNA binding motif protein 5 |
| chr17_-_17872573 | 0.10 |
ENSRNOT00000082733
|
Rnf144b
|
ring finger protein 144B |
| chr17_-_32783427 | 0.10 |
ENSRNOT00000059921
|
Serpinb6b
|
serine (or cysteine) peptidase inhibitor, clade B, member 6b |
| chr13_+_68785827 | 0.10 |
ENSRNOT00000003517
|
Trmt1l
|
tRNA methyltransferase 1-like |
| chr13_-_80738634 | 0.10 |
ENSRNOT00000081551
|
Fmo1
|
flavin containing monooxygenase 1 |
| chr8_-_40137390 | 0.10 |
ENSRNOT00000042717
|
Panx3
|
pannexin 3 |
| chr4_-_147505838 | 0.10 |
ENSRNOT00000086570
|
Mkrn2os
|
MKRN2 opposite strand |
| chr5_-_113532878 | 0.09 |
ENSRNOT00000010173
|
Caap1
|
caspase activity and apoptosis inhibitor 1 |
| chr10_-_34990943 | 0.09 |
ENSRNOT00000075555
|
Rmnd5b
|
required for meiotic nuclear division 5 homolog B |
| chr4_-_126071261 | 0.09 |
ENSRNOT00000080234
|
LOC100361920
|
dynein light chain 1-like |
| chr2_+_185343883 | 0.09 |
ENSRNOT00000093640
|
Sh3d19
|
SH3 domain containing 19 |
| chr9_-_121725716 | 0.09 |
ENSRNOT00000087405
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chrX_+_68891227 | 0.09 |
ENSRNOT00000009635
|
Efnb1
|
ephrin B1 |
| chr9_-_30844199 | 0.09 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr8_-_45137893 | 0.09 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr2_+_189454340 | 0.09 |
ENSRNOT00000087063
|
Nup210l
|
nucleoporin 210-like |
| chr20_+_10438444 | 0.09 |
ENSRNOT00000071248
ENSRNOT00000075545 |
Cryaa
|
crystallin, alpha A |
| chr13_-_91228901 | 0.09 |
ENSRNOT00000071728
ENSRNOT00000073643 ENSRNOT00000071897 |
LOC100911825
|
low affinity immunoglobulin gamma Fc region receptor III-like |
| chr9_+_14560504 | 0.09 |
ENSRNOT00000091532
|
Nfya
|
nuclear transcription factor Y subunit alpha |
| chr10_-_90151042 | 0.09 |
ENSRNOT00000055187
|
Hdac5
|
histone deacetylase 5 |
| chr12_+_10255416 | 0.09 |
ENSRNOT00000092720
|
Gpr12
|
G protein-coupled receptor 12 |
| chr12_-_46920952 | 0.09 |
ENSRNOT00000001532
|
Msi1
|
musashi RNA-binding protein 1 |
| chr2_+_198721724 | 0.09 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr12_-_17322608 | 0.09 |
ENSRNOT00000033038
|
LOC102546864
|
uncharacterized LOC102546864 |
| chr3_-_43119159 | 0.09 |
ENSRNOT00000041394
|
Nr4a2
|
nuclear receptor subfamily 4, group A, member 2 |
| chr11_+_43143882 | 0.09 |
ENSRNOT00000041445
|
Olr1528
|
olfactory receptor 1528 |
| chr16_-_75481115 | 0.09 |
ENSRNOT00000035128
|
Defa7
|
defensin alpha 7 |
| chr4_-_165541314 | 0.09 |
ENSRNOT00000013833
|
Styk1
|
serine/threonine/tyrosine kinase 1 |
| chr4_+_5644260 | 0.09 |
ENSRNOT00000041876
|
Actr3b
|
ARP3 actin related protein 3 homolog B |
| chr4_-_113954272 | 0.09 |
ENSRNOT00000039966
ENSRNOT00000082996 |
LOC103692173
|
WW domain-binding protein 1 |
| chr7_-_76294663 | 0.09 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr1_+_88113445 | 0.09 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr9_-_103207190 | 0.09 |
ENSRNOT00000026010
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr1_-_81450094 | 0.09 |
ENSRNOT00000035356
|
Zfp575
|
zinc finger protein 575 |
| chr6_-_115513354 | 0.09 |
ENSRNOT00000005881
|
Ston2
|
stonin 2 |
| chr11_+_37798370 | 0.09 |
ENSRNOT00000002679
|
Bace2
|
beta-site APP-cleaving enzyme 2 |
| chr18_+_30581530 | 0.08 |
ENSRNOT00000048166
|
Pcdhb20
|
protocadherin beta 20 |
| chr20_-_4561062 | 0.08 |
ENSRNOT00000065044
ENSRNOT00000092698 ENSRNOT00000060607 |
Cfb
C2
|
complement factor B complement C2 |
| chr18_-_29611212 | 0.08 |
ENSRNOT00000022685
|
Dnd1
|
DND microRNA-mediated repression inhibitor 1 |
| chr16_+_20521956 | 0.08 |
ENSRNOT00000026597
|
Pgpep1
|
pyroglutamyl-peptidase I |
| chr13_-_110784209 | 0.08 |
ENSRNOT00000071698
|
Traf5
|
TNF receptor-associated factor 5 |
| chr7_-_120380200 | 0.08 |
ENSRNOT00000014878
|
RGD1359634
|
similar to RIKEN cDNA 1700088E04 |
| chr1_-_82003691 | 0.08 |
ENSRNOT00000084569
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr10_-_75202030 | 0.08 |
ENSRNOT00000050349
|
Olr1522
|
olfactory receptor 1522 |
| chrX_-_158261717 | 0.08 |
ENSRNOT00000086804
|
RGD1561558
|
similar to RIKEN cDNA 1700001F22 |
| chr7_-_29986163 | 0.08 |
ENSRNOT00000033123
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr5_-_164927869 | 0.08 |
ENSRNOT00000012080
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr5_-_155916893 | 0.08 |
ENSRNOT00000055947
|
Ldlrad2
|
low density lipoprotein receptor class A domain containing 2 |
| chr10_+_72197977 | 0.08 |
ENSRNOT00000003886
|
Myo19
|
myosin XIX |
| chr19_-_25378974 | 0.08 |
ENSRNOT00000011150
|
Ccdc130
|
coiled-coil domain containing 130 |
| chr11_+_57108956 | 0.08 |
ENSRNOT00000035485
|
Cd96
|
CD96 molecule |
| chr1_-_169513537 | 0.08 |
ENSRNOT00000078058
|
Trim30c
|
tripartite motif-containing 30C |
| chr13_-_89661150 | 0.08 |
ENSRNOT00000058390
|
Usp21
|
ubiquitin specific peptidase 21 |
| chr4_-_114819848 | 0.08 |
ENSRNOT00000084381
|
Wbp1
|
WW domain binding protein 1 |
| chr7_+_140829076 | 0.08 |
ENSRNOT00000086179
|
Spats2
|
spermatogenesis associated, serine-rich 2 |
| chr10_-_34361212 | 0.08 |
ENSRNOT00000072557
|
Olr1385
|
olfactory receptor 1385 |
| chr7_+_140464999 | 0.08 |
ENSRNOT00000090156
|
Wnt1
|
wingless-type MMTV integration site family, member 1 |
| chr14_-_22890958 | 0.08 |
ENSRNOT00000031916
|
LOC100911104
|
lymphocyte antigen 6B-like |
| chr12_+_13323547 | 0.08 |
ENSRNOT00000074138
|
Zfp853
|
zinc finger protein 853 |
| chr2_-_257864385 | 0.08 |
ENSRNOT00000072048
|
Ak5
|
adenylate kinase 5 |
| chr11_+_86903122 | 0.08 |
ENSRNOT00000048063
|
Zdhhc8
|
zinc finger, DHHC-type containing 8 |
| chr7_+_12542713 | 0.08 |
ENSRNOT00000080089
|
Med16
|
mediator complex subunit 16 |
| chr7_-_2712723 | 0.08 |
ENSRNOT00000004363
|
Il23a
|
interleukin 23 subunit alpha |
| chr17_-_10364503 | 0.08 |
ENSRNOT00000086380
ENSRNOT00000041709 |
Tspan17
|
tetraspanin 17 |
| chr13_-_89433815 | 0.08 |
ENSRNOT00000091541
|
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr9_+_95202632 | 0.08 |
ENSRNOT00000025652
|
Ugt1a5
|
UDP glucuronosyltransferase family 1 member A5 |
| chr13_-_61591139 | 0.08 |
ENSRNOT00000005324
|
Rgs18
|
regulator of G-protein signaling 18 |
| chr8_-_119889661 | 0.08 |
ENSRNOT00000011780
|
Stac
|
SH3 and cysteine rich domain |
| chr18_+_44716226 | 0.08 |
ENSRNOT00000086431
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr12_+_46989951 | 0.08 |
ENSRNOT00000001536
|
Gatc
|
glutamyl-tRNA amidotransferase subunit C |
| chr1_-_241155537 | 0.08 |
ENSRNOT00000034216
ENSRNOT00000073493 |
Mamdc2
|
MAM domain containing 2 |
| chr13_-_90977734 | 0.08 |
ENSRNOT00000011869
|
Slamf8
|
SLAM family member 8 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx2_Bcl11a
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.1 | 0.4 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 0.3 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.1 | 0.3 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.2 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
| 0.1 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.2 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.2 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.0 | 0.2 | GO:0032910 | transforming growth factor beta3 production(GO:0032907) regulation of transforming growth factor beta3 production(GO:0032910) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:0060838 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
| 0.0 | 0.2 | GO:0070366 | regulation of hepatocyte differentiation(GO:0070366) |
| 0.0 | 0.3 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.3 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.0 | GO:0002536 | respiratory burst involved in inflammatory response(GO:0002536) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.1 | GO:2001274 | negative regulation of glucose import in response to insulin stimulus(GO:2001274) |
| 0.0 | 0.1 | GO:0046951 | ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.1 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.2 | GO:2001204 | regulation of osteoclast development(GO:2001204) |
| 0.0 | 0.1 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.0 | 0.1 | GO:1904954 | Spemann organizer formation(GO:0060061) dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) positive regulation of dermatome development(GO:0061184) Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904953) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.0 | 0.2 | GO:0052697 | xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:2000328 | regulation of NK T cell proliferation(GO:0051140) positive regulation of NK T cell proliferation(GO:0051142) regulation of T-helper 17 cell lineage commitment(GO:2000328) |
| 0.0 | 0.1 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:0002232 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) |
| 0.0 | 0.2 | GO:0021612 | facial nerve structural organization(GO:0021612) |
| 0.0 | 0.1 | GO:0038027 | apolipoprotein A-I-mediated signaling pathway(GO:0038027) |
| 0.0 | 0.1 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:2000427 | positive regulation of apoptotic cell clearance(GO:2000427) |
| 0.0 | 0.1 | GO:0042985 | negative regulation of amyloid precursor protein biosynthetic process(GO:0042985) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.0 | GO:0098758 | response to interleukin-8(GO:0098758) cellular response to interleukin-8(GO:0098759) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.4 | GO:0060218 | hematopoietic stem cell differentiation(GO:0060218) |
| 0.0 | 0.3 | GO:0043653 | mitochondrial fragmentation involved in apoptotic process(GO:0043653) |
| 0.0 | 0.2 | GO:1900017 | positive regulation of cytokine production involved in inflammatory response(GO:1900017) |
| 0.0 | 0.0 | GO:0002277 | myeloid dendritic cell activation involved in immune response(GO:0002277) |
| 0.0 | 0.1 | GO:0070309 | lens fiber cell morphogenesis(GO:0070309) |
| 0.0 | 0.1 | GO:0036265 | RNA (guanine-N7)-methylation(GO:0036265) |
| 0.0 | 0.1 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 0.1 | GO:0002331 | pre-B cell allelic exclusion(GO:0002331) |
| 0.0 | 0.1 | GO:0051866 | general adaptation syndrome(GO:0051866) |
| 0.0 | 0.0 | GO:0051311 | meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
| 0.0 | 0.1 | GO:0001788 | antibody-dependent cellular cytotoxicity(GO:0001788) |
| 0.0 | 0.1 | GO:0097394 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.1 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.3 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.0 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.2 | GO:0090026 | positive regulation of monocyte chemotaxis(GO:0090026) |
| 0.0 | 0.1 | GO:0010808 | positive regulation of synaptic vesicle priming(GO:0010808) presynaptic dense core granule exocytosis(GO:0099525) |
| 0.0 | 0.3 | GO:0007190 | activation of adenylate cyclase activity(GO:0007190) |
| 0.0 | 0.0 | GO:0006742 | NADP catabolic process(GO:0006742) |
| 0.0 | 0.0 | GO:0002465 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.1 | GO:1902019 | regulation of cilium movement involved in cell motility(GO:0060295) regulation of cilium beat frequency involved in ciliary motility(GO:0060296) regulation of cilium-dependent cell motility(GO:1902019) |
| 0.0 | 0.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.1 | GO:0010572 | positive regulation of platelet activation(GO:0010572) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 0.2 | GO:0038094 | Fc-gamma receptor signaling pathway(GO:0038094) |
| 0.0 | 0.1 | GO:0060161 | peptidyl-arginine methylation, to symmetrical-dimethyl arginine(GO:0019918) positive regulation of dopamine receptor signaling pathway(GO:0060161) |
| 0.0 | 0.1 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.0 | 0.6 | GO:0045494 | photoreceptor cell maintenance(GO:0045494) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:1904640 | glycerol biosynthetic process(GO:0006114) response to methionine(GO:1904640) |
| 0.0 | 0.1 | GO:0030205 | dermatan sulfate metabolic process(GO:0030205) |
| 0.0 | 0.1 | GO:0010759 | positive regulation of macrophage chemotaxis(GO:0010759) |
| 0.0 | 0.2 | GO:0002283 | neutrophil activation involved in immune response(GO:0002283) |
| 0.0 | 0.1 | GO:0048304 | positive regulation of isotype switching to IgG isotypes(GO:0048304) |
| 0.0 | 0.0 | GO:0060298 | positive regulation of sarcomere organization(GO:0060298) |
| 0.0 | 0.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.2 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
| 0.0 | 0.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:1902255 | positive regulation of intrinsic apoptotic signaling pathway by p53 class mediator(GO:1902255) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.0 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0002727 | natural killer cell cytokine production(GO:0002370) regulation of natural killer cell cytokine production(GO:0002727) |
| 0.0 | 0.0 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.0 | GO:1900224 | zygotic specification of dorsal/ventral axis(GO:0007352) positive regulation of nodal signaling pathway involved in determination of lateral mesoderm left/right asymmetry(GO:1900224) |
| 0.0 | 0.1 | GO:0090086 | negative regulation of protein deubiquitination(GO:0090086) |
| 0.0 | 0.1 | GO:0048368 | lateral mesoderm development(GO:0048368) |
| 0.0 | 0.1 | GO:0034627 | 'de novo' NAD biosynthetic process(GO:0034627) |
| 0.0 | 0.0 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0009730 | detection of carbohydrate stimulus(GO:0009730) detection of hexose stimulus(GO:0009732) detection of monosaccharide stimulus(GO:0034287) detection of glucose(GO:0051594) |
| 0.0 | 0.0 | GO:0003402 | planar cell polarity pathway involved in axis elongation(GO:0003402) |
| 0.0 | 0.1 | GO:0060965 | negative regulation of gene silencing by miRNA(GO:0060965) |
| 0.0 | 0.1 | GO:0060481 | lobar bronchus epithelium development(GO:0060481) |
| 0.0 | 0.0 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.0 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.0 | GO:0090289 | regulation of osteoclast proliferation(GO:0090289) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.1 | 0.2 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:0034774 | secretory granule lumen(GO:0034774) |
| 0.0 | 0.6 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.2 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.2 | GO:0031501 | mannosyltransferase complex(GO:0031501) |
| 0.0 | 0.2 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.4 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.4 | GO:0032009 | early phagosome(GO:0032009) |
| 0.0 | 0.1 | GO:0097169 | IPAF inflammasome complex(GO:0072557) NLRP1 inflammasome complex(GO:0072558) AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.2 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.1 | GO:1990707 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.0 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.0 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.4 | GO:0000159 | protein phosphatase type 2A complex(GO:0000159) |
| 0.0 | 0.1 | GO:0016602 | CCAAT-binding factor complex(GO:0016602) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.0 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.0 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.1 | 0.4 | GO:0004999 | vasoactive intestinal polypeptide receptor activity(GO:0004999) |
| 0.1 | 0.2 | GO:0071791 | chemokine (C-C motif) ligand 5 binding(GO:0071791) |
| 0.0 | 0.1 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:0030172 | troponin C binding(GO:0030172) |
| 0.0 | 0.2 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.2 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0004809 | tRNA (guanine-N2-)-methyltransferase activity(GO:0004809) |
| 0.0 | 0.4 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.0 | 0.1 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.2 | GO:0022851 | GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.2 | GO:0030274 | LIM domain binding(GO:0030274) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.3 | GO:0035325 | Toll-like receptor binding(GO:0035325) |
| 0.0 | 0.2 | GO:0030368 | interleukin-17 receptor activity(GO:0030368) |
| 0.0 | 0.1 | GO:0004046 | aminoacylase activity(GO:0004046) |
| 0.0 | 0.1 | GO:0102345 | 3-hydroxy-behenoyl-CoA dehydratase activity(GO:0102344) 3-hydroxy-lignoceroyl-CoA dehydratase activity(GO:0102345) |
| 0.0 | 0.1 | GO:0016623 | aldehyde oxidase activity(GO:0004031) oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.2 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.0 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.0 | GO:0008413 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.1 | GO:0034186 | apolipoprotein A-I binding(GO:0034186) |
| 0.0 | 0.1 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0052813 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.0 | 0.1 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.0 | GO:0004918 | interleukin-8 receptor activity(GO:0004918) |
| 0.0 | 0.1 | GO:0035243 | protein-arginine omega-N symmetric methyltransferase activity(GO:0035243) |
| 0.0 | 0.1 | GO:0015245 | fatty acid transporter activity(GO:0015245) very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.1 | GO:0016884 | carbon-nitrogen ligase activity, with glutamine as amido-N-donor(GO:0016884) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0019158 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.0 | GO:0030337 | DNA polymerase processivity factor activity(GO:0030337) |
| 0.0 | 0.2 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.0 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0019992 | diacylglycerol binding(GO:0019992) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |
| 0.0 | 0.0 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.0 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0004983 | neuropeptide Y receptor activity(GO:0004983) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.0 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.0 | 0.3 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.9 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 0.0 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |
| 0.1 | 0.2 | REACTOME DEFENSINS | Genes involved in Defensins |
| 0.0 | 0.5 | REACTOME SIGNAL REGULATORY PROTEIN SIRP FAMILY INTERACTIONS | Genes involved in Signal regulatory protein (SIRP) family interactions |
| 0.0 | 0.2 | REACTOME SIGNALING BY FGFR | Genes involved in Signaling by FGFR |
| 0.0 | 0.1 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME TRAFFICKING AND PROCESSING OF ENDOSOMAL TLR | Genes involved in Trafficking and processing of endosomal TLR |
| 0.0 | 0.4 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF BILE ACIDS AND BILE SALTS VIA 24 HYDROXYCHOLESTEROL | Genes involved in Synthesis of bile acids and bile salts via 24-hydroxycholesterol |
| 0.0 | 0.1 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.5 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 0.1 | REACTOME CD28 CO STIMULATION | Genes involved in CD28 co-stimulation |
| 0.0 | 0.3 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |