Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
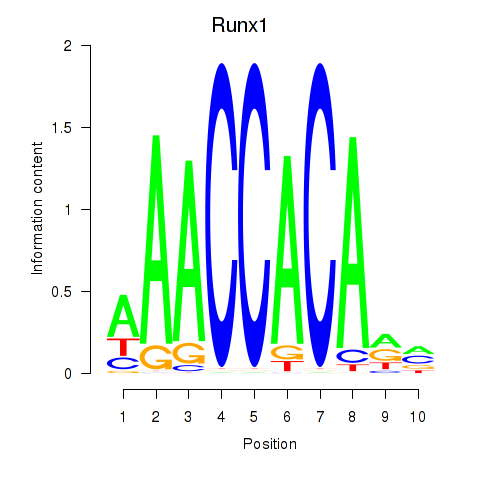
Results for Runx1
Z-value: 0.50
Transcription factors associated with Runx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Runx1
|
ENSRNOG00000001704 | runt-related transcription factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Runx1 | rn6_v1_chr11_-_33003021_33003021 | 0.48 | 4.1e-01 | Click! |
Activity profile of Runx1 motif
Sorted Z-values of Runx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_187742747 | 0.33 |
ENSRNOT00000026530
|
Bglap
|
bone gamma-carboxyglutamate protein |
| chr10_-_85435016 | 0.28 |
ENSRNOT00000079921
|
4933428G20Rik
|
RIKEN cDNA 4933428G20 gene |
| chr14_-_78902063 | 0.18 |
ENSRNOT00000088469
|
Ppp2r2c
|
protein phosphatase 2, regulatory subunit B, gamma |
| chr1_+_44311513 | 0.17 |
ENSRNOT00000065386
|
Tiam2
|
T-cell lymphoma invasion and metastasis 2 |
| chr18_-_55916220 | 0.17 |
ENSRNOT00000025934
|
Synpo
|
synaptopodin |
| chr20_-_22459025 | 0.16 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr10_-_38985466 | 0.15 |
ENSRNOT00000010121
|
Il13
|
interleukin 13 |
| chr13_-_83202864 | 0.13 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr9_-_15700235 | 0.13 |
ENSRNOT00000088713
ENSRNOT00000035907 |
Trerf1
|
transcriptional regulating factor 1 |
| chr1_+_100577056 | 0.12 |
ENSRNOT00000026992
|
Napsa
|
napsin A aspartic peptidase |
| chr4_-_117126822 | 0.12 |
ENSRNOT00000086720
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr10_+_104582955 | 0.11 |
ENSRNOT00000009733
|
Unk
|
unkempt family zinc finger |
| chr1_+_190666149 | 0.11 |
ENSRNOT00000089361
|
LOC102547219
|
uncharacterized LOC102547219 |
| chr7_-_145450233 | 0.11 |
ENSRNOT00000092974
ENSRNOT00000021523 |
Calcoco1
|
calcium binding and coiled coil domain 1 |
| chr16_+_46731403 | 0.11 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr1_-_226887156 | 0.11 |
ENSRNOT00000054809
ENSRNOT00000028347 |
Cd6
|
Cd6 molecule |
| chrX_-_159841072 | 0.10 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr1_-_82004538 | 0.10 |
ENSRNOT00000087572
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr14_-_6679878 | 0.10 |
ENSRNOT00000075989
ENSRNOT00000067875 |
Spp1
|
secreted phosphoprotein 1 |
| chr14_-_81399353 | 0.10 |
ENSRNOT00000018340
|
Add1
|
adducin 1 |
| chr1_-_82003691 | 0.10 |
ENSRNOT00000084569
|
Pou2f2
|
POU class 2 homeobox 2 |
| chr9_+_118849302 | 0.10 |
ENSRNOT00000087592
|
Dlgap1
|
DLG associated protein 1 |
| chr5_+_151362019 | 0.09 |
ENSRNOT00000077083
ENSRNOT00000067801 |
Wasf2
|
WAS protein family, member 2 |
| chr6_+_52631092 | 0.09 |
ENSRNOT00000014054
|
Atxn7l1
|
ataxin 7-like 1 |
| chr11_-_33003021 | 0.09 |
ENSRNOT00000084134
|
Runx1
|
runt-related transcription factor 1 |
| chr13_+_70379346 | 0.09 |
ENSRNOT00000038183
|
Nmnat2
|
nicotinamide nucleotide adenylyltransferase 2 |
| chr8_-_62987182 | 0.09 |
ENSRNOT00000070885
|
Islr2
|
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr4_-_40036563 | 0.09 |
ENSRNOT00000084531
|
Tmem168
|
transmembrane protein 168 |
| chr9_+_18564927 | 0.09 |
ENSRNOT00000061014
|
Runx2
|
runt-related transcription factor 2 |
| chr5_+_25725683 | 0.08 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr4_-_155275161 | 0.08 |
ENSRNOT00000032690
|
Rimklb
|
ribosomal modification protein rimK-like family member B |
| chr9_+_71247781 | 0.07 |
ENSRNOT00000049654
|
Creb1
|
cAMP responsive element binding protein 1 |
| chr8_-_47339343 | 0.07 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr14_-_55081551 | 0.07 |
ENSRNOT00000049245
|
Pcdh7
|
protocadherin 7 |
| chr10_-_83655182 | 0.06 |
ENSRNOT00000007897
|
Abi3
|
ABI family, member 3 |
| chr1_+_47032113 | 0.06 |
ENSRNOT00000024412
|
Tulp4
|
tubby like protein 4 |
| chr10_+_75087892 | 0.06 |
ENSRNOT00000065910
|
Mpo
|
myeloperoxidase |
| chr8_-_96266342 | 0.06 |
ENSRNOT00000078891
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr20_-_4508197 | 0.06 |
ENSRNOT00000086027
ENSRNOT00000000514 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr2_+_116032455 | 0.06 |
ENSRNOT00000066495
|
Phc3
|
polyhomeotic homolog 3 |
| chr5_+_25253010 | 0.05 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr8_-_96132634 | 0.05 |
ENSRNOT00000041655
|
Syncrip
|
synaptotagmin binding, cytoplasmic RNA interacting protein |
| chr14_+_86828355 | 0.05 |
ENSRNOT00000088611
|
Ccm2
|
CCM2 scaffolding protein |
| chr10_-_65200109 | 0.05 |
ENSRNOT00000030501
|
Nufip2
|
NUFIP2, FMR1 interacting protein 2 |
| chr20_-_2678141 | 0.05 |
ENSRNOT00000072377
ENSRNOT00000083833 |
C4a
|
complement component 4A (Rodgers blood group) |
| chr20_-_4316715 | 0.05 |
ENSRNOT00000031704
|
C4b
|
complement C4B (Chido blood group) |
| chr16_+_83824430 | 0.05 |
ENSRNOT00000032918
|
Irs2
|
insulin receptor substrate 2 |
| chr2_-_210550490 | 0.05 |
ENSRNOT00000081835
ENSRNOT00000025222 ENSRNOT00000086403 |
Csf1
|
colony stimulating factor 1 |
| chr9_+_67546408 | 0.05 |
ENSRNOT00000013701
|
Cd28
|
Cd28 molecule |
| chr2_+_143475323 | 0.04 |
ENSRNOT00000044028
ENSRNOT00000015437 |
Trpc4
|
transient receptor potential cation channel, subfamily C, member 4 |
| chr4_+_65637407 | 0.04 |
ENSRNOT00000047954
|
Trim24
|
tripartite motif-containing 24 |
| chr1_-_219544328 | 0.04 |
ENSRNOT00000025847
|
Grk2
|
G protein-coupled receptor kinase 2 |
| chr2_-_44981458 | 0.04 |
ENSRNOT00000014134
|
Gzma
|
granzyme A |
| chr17_-_87826421 | 0.04 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr3_-_44177689 | 0.04 |
ENSRNOT00000006387
|
Cytip
|
cytohesin 1 interacting protein |
| chr1_+_79982639 | 0.04 |
ENSRNOT00000071916
|
Dmwd
|
dystrophia myotonica, WD repeat containing |
| chr1_+_5240180 | 0.03 |
ENSRNOT00000019893
ENSRNOT00000088292 |
Shprh
|
SNF2 histone linker PHD RING helicase |
| chr3_+_66673071 | 0.03 |
ENSRNOT00000034769
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr2_+_205553163 | 0.03 |
ENSRNOT00000039572
|
Nras
|
neuroblastoma RAS viral oncogene homolog |
| chr3_+_66673246 | 0.03 |
ENSRNOT00000081338
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr9_-_81864202 | 0.03 |
ENSRNOT00000084964
|
Zfp142
|
zinc finger protein 142 |
| chr15_-_42947656 | 0.03 |
ENSRNOT00000030007
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr10_+_86950557 | 0.03 |
ENSRNOT00000014153
|
Igfbp4
|
insulin-like growth factor binding protein 4 |
| chr2_-_203413124 | 0.03 |
ENSRNOT00000077898
ENSRNOT00000056140 |
Cd101
|
CD101 molecule |
| chr1_+_167522895 | 0.02 |
ENSRNOT00000092987
|
Stim1
|
stromal interaction molecule 1 |
| chr15_-_27716008 | 0.02 |
ENSRNOT00000011741
|
Ttc5
|
tetratricopeptide repeat domain 5 |
| chr3_+_104816987 | 0.02 |
ENSRNOT00000042103
ENSRNOT00000044625 |
Fmn1
|
formin 1 |
| chr8_-_40137390 | 0.02 |
ENSRNOT00000042717
|
Panx3
|
pannexin 3 |
| chr1_+_192233910 | 0.02 |
ENSRNOT00000016418
ENSRNOT00000016442 |
Prkcb
|
protein kinase C, beta |
| chr7_+_140464999 | 0.02 |
ENSRNOT00000090156
|
Wnt1
|
wingless-type MMTV integration site family, member 1 |
| chr1_-_87825333 | 0.02 |
ENSRNOT00000072689
|
Zfp74
|
zinc finger protein 74 |
| chr7_-_76294663 | 0.01 |
ENSRNOT00000064513
|
Ncald
|
neurocalcin delta |
| chr11_+_88122271 | 0.01 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr11_+_44001579 | 0.01 |
ENSRNOT00000002258
|
Gpr15
|
G protein-coupled receptor 15 |
| chr3_-_146491837 | 0.01 |
ENSRNOT00000061469
|
Vsx1
|
visual system homeobox 1 |
| chr19_+_10024947 | 0.01 |
ENSRNOT00000061392
|
Cfap20
|
cilia and flagella associated protein 20 |
| chr7_-_62162453 | 0.01 |
ENSRNOT00000010720
|
Cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr1_-_88240057 | 0.01 |
ENSRNOT00000073092
|
LOC103690038
|
zinc finger protein 569 |
| chr3_+_170399302 | 0.01 |
ENSRNOT00000035245
|
Cass4
|
Cas scaffolding protein family member 4 |
| chr3_+_79729739 | 0.01 |
ENSRNOT00000084833
|
Kbtbd4
|
kelch repeat and BTB domain containing 4 |
| chr4_-_184096806 | 0.00 |
ENSRNOT00000055433
|
LOC100362344
|
mKIAA1238 protein-like |
| chr7_+_121930615 | 0.00 |
ENSRNOT00000091270
ENSRNOT00000033975 |
Tnrc6b
|
trinucleotide repeat containing 6B |
| chr6_+_109562587 | 0.00 |
ENSRNOT00000011563
|
Batf
|
basic leucine zipper ATF-like transcription factor |
| chr7_-_144880092 | 0.00 |
ENSRNOT00000055281
|
Nfe2
|
nuclear factor, erythroid 2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Runx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0002380 | immunoglobulin secretion involved in immune response(GO:0002380) |
| 0.1 | 0.2 | GO:2000538 | regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.2 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.2 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.0 | 0.3 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:2000872 | positive regulation of progesterone secretion(GO:2000872) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0070164 | negative regulation of adiponectin secretion(GO:0070164) |
| 0.0 | 0.1 | GO:0032916 | positive regulation of transforming growth factor beta3 production(GO:0032916) |
| 0.0 | 0.1 | GO:0002149 | hypochlorous acid metabolic process(GO:0002148) hypochlorous acid biosynthetic process(GO:0002149) |
| 0.0 | 0.1 | GO:0090367 | negative regulation of mRNA modification(GO:0090367) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.0 | GO:1904139 | regulation of macrophage colony-stimulating factor signaling pathway(GO:1902226) regulation of response to macrophage colony-stimulating factor(GO:1903969) regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903972) microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.0 | GO:0010748 | negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.2 | GO:0070262 | peptidyl-serine dephosphorylation(GO:0070262) |
| 0.0 | 0.1 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0089717 | spanning component of plasma membrane(GO:0044214) spanning component of membrane(GO:0089717) |
| 0.0 | 0.1 | GO:1990589 | ATF4-CREB1 transcription factor complex(GO:1990589) |
| 0.0 | 0.0 | GO:1990682 | CSF1-CSF1R complex(GO:1990682) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0031209 | SCAR complex(GO:0031209) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004515 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.1 | GO:1990715 | mRNA CDS binding(GO:1990715) |
| 0.0 | 0.0 | GO:0005157 | macrophage colony-stimulating factor receptor binding(GO:0005157) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |