Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
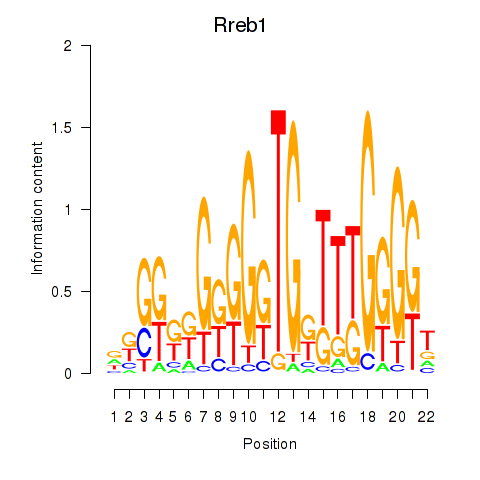
Results for Rreb1
Z-value: 0.61
Transcription factors associated with Rreb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rreb1
|
ENSRNOG00000015701 | ras responsive element binding protein 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rreb1 | rn6_v1_chr17_-_27602934_27602934 | 0.74 | 1.5e-01 | Click! |
Activity profile of Rreb1 motif
Sorted Z-values of Rreb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_100577056 | 0.82 |
ENSRNOT00000026992
|
Napsa
|
napsin A aspartic peptidase |
| chr5_-_100647727 | 0.21 |
ENSRNOT00000067435
|
Nfib
|
nuclear factor I/B |
| chr4_+_88834066 | 0.21 |
ENSRNOT00000009546
|
Abcg2
|
ATP-binding cassette, subfamily G (WHITE), member 2 |
| chr12_-_37574750 | 0.20 |
ENSRNOT00000066253
|
Kmt5a
|
lysine methyltransferase 5A |
| chr17_-_32661865 | 0.17 |
ENSRNOT00000022194
|
Serpinb9
|
serpin family B member 9 |
| chr8_-_129919120 | 0.17 |
ENSRNOT00000074423
|
Ulk4
|
unc-51 like kinase 4 |
| chr1_-_42486035 | 0.17 |
ENSRNOT00000025398
|
Mtrf1l
|
mitochondrial translational release factor 1-like |
| chr20_+_4363508 | 0.17 |
ENSRNOT00000077205
|
Ager
|
advanced glycosylation end product-specific receptor |
| chr2_-_22798214 | 0.17 |
ENSRNOT00000016135
|
Papd4
|
poly(A) RNA polymerase D4, non-canonical |
| chr12_-_25638797 | 0.16 |
ENSRNOT00000002033
|
Gatsl2
|
GATS protein-like 2 |
| chr12_+_38144855 | 0.15 |
ENSRNOT00000032274
|
Hcar1
|
hydroxycarboxylic acid receptor 1 |
| chr14_-_92577936 | 0.15 |
ENSRNOT00000086154
|
Cobl
|
cordon-bleu WH2 repeat protein |
| chr18_+_15467870 | 0.14 |
ENSRNOT00000091696
|
B4galt6
|
beta-1,4-galactosyltransferase 6 |
| chr2_-_207300854 | 0.14 |
ENSRNOT00000018061
|
Mov10
|
Mov10 RISC complex RNA helicase |
| chr15_-_84748525 | 0.14 |
ENSRNOT00000090009
|
Klf12
|
Kruppel-like factor 12 |
| chr10_-_90030423 | 0.14 |
ENSRNOT00000092150
|
Mpp2
|
membrane palmitoylated protein 2 |
| chr3_-_93734282 | 0.14 |
ENSRNOT00000012428
|
Caprin1
|
cell cycle associated protein 1 |
| chr5_-_7874909 | 0.14 |
ENSRNOT00000064774
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr4_-_169999873 | 0.13 |
ENSRNOT00000011697
|
Grin2b
|
glutamate ionotropic receptor NMDA type subunit 2B |
| chr1_+_77782825 | 0.13 |
ENSRNOT00000058230
|
AABR07002623.1
|
|
| chr2_+_210045161 | 0.13 |
ENSRNOT00000024455
|
Slc16a4
|
solute carrier family 16, member 4 |
| chr14_+_115166416 | 0.13 |
ENSRNOT00000088916
ENSRNOT00000078329 |
Psme4
|
proteasome activator subunit 4 |
| chr7_-_130128589 | 0.13 |
ENSRNOT00000079777
ENSRNOT00000009325 |
Mapk11
|
mitogen-activated protein kinase 11 |
| chr2_+_198303168 | 0.12 |
ENSRNOT00000056262
|
Mtmr11
|
myotubularin related protein 11 |
| chr1_-_140262452 | 0.12 |
ENSRNOT00000046849
ENSRNOT00000045165 ENSRNOT00000025536 ENSRNOT00000041839 |
Ntrk3
|
neurotrophic receptor tyrosine kinase 3 |
| chr3_-_162579201 | 0.12 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr3_-_29996865 | 0.12 |
ENSRNOT00000080382
|
Zeb2
|
zinc finger E-box binding homeobox 2 |
| chr10_-_74298599 | 0.12 |
ENSRNOT00000007379
|
Ypel2
|
yippee-like 2 |
| chr13_-_51955735 | 0.12 |
ENSRNOT00000007597
|
Ptprv
|
protein tyrosine phosphatase, receptor type, V |
| chr2_+_86891092 | 0.12 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr16_+_31734944 | 0.11 |
ENSRNOT00000059673
|
Palld
|
palladin, cytoskeletal associated protein |
| chr1_-_19376301 | 0.11 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr2_+_23289374 | 0.11 |
ENSRNOT00000090666
ENSRNOT00000032783 |
Dmgdh
|
dimethylglycine dehydrogenase |
| chr12_+_40695756 | 0.11 |
ENSRNOT00000091375
|
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr12_+_40695520 | 0.11 |
ENSRNOT00000033984
|
Trafd1
|
TRAF type zinc finger domain containing 1 |
| chr18_-_399242 | 0.11 |
ENSRNOT00000045926
|
F8
|
coagulation factor VIII |
| chr9_-_27761365 | 0.11 |
ENSRNOT00000018552
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chr1_+_266380973 | 0.11 |
ENSRNOT00000080509
|
Wbp1l
|
WW domain binding protein 1-like |
| chr12_+_25498198 | 0.11 |
ENSRNOT00000076916
|
Ncf1
|
neutrophil cytosolic factor 1 |
| chr1_-_101323960 | 0.10 |
ENSRNOT00000041513
|
Trpm4
|
transient receptor potential cation channel, subfamily M, member 4 |
| chr2_-_23289266 | 0.10 |
ENSRNOT00000061708
|
Bhmt2
|
betaine-homocysteine S-methyltransferase 2 |
| chrX_+_71155601 | 0.10 |
ENSRNOT00000076453
ENSRNOT00000048521 |
Foxo4
|
forkhead box O4 |
| chr18_+_24540659 | 0.10 |
ENSRNOT00000061074
|
Ammecr1l
|
AMMECR1 like |
| chr17_-_17872573 | 0.10 |
ENSRNOT00000082733
|
Rnf144b
|
ring finger protein 144B |
| chr2_+_235596907 | 0.10 |
ENSRNOT00000071463
ENSRNOT00000075728 |
Col25a1
|
collagen type XXV alpha 1 chain |
| chr8_+_44047592 | 0.10 |
ENSRNOT00000085084
|
Zfp202
|
zinc finger protein 202 |
| chr7_-_12275609 | 0.10 |
ENSRNOT00000086061
|
Apc2
|
APC2, WNT signaling pathway regulator |
| chrX_-_152641679 | 0.10 |
ENSRNOT00000080277
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr5_+_147476221 | 0.09 |
ENSRNOT00000010730
|
Sync
|
syncoilin, intermediate filament protein |
| chr12_-_46494174 | 0.09 |
ENSRNOT00000086716
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr3_+_3310954 | 0.09 |
ENSRNOT00000061773
|
Kcnt1
|
potassium sodium-activated channel subfamily T member 1 |
| chr4_-_148446303 | 0.09 |
ENSRNOT00000017633
|
Alox5
|
arachidonate 5-lipoxygenase |
| chr20_-_30748784 | 0.09 |
ENSRNOT00000084391
|
Sgpl1
|
sphingosine-1-phosphate lyase 1 |
| chr10_+_104320743 | 0.09 |
ENSRNOT00000005684
|
Tmem94
|
transmembrane protein 94 |
| chr15_+_83495377 | 0.09 |
ENSRNOT00000036350
|
Pibf1
|
progesterone immunomodulatory binding factor 1 |
| chr14_+_36689096 | 0.09 |
ENSRNOT00000083100
|
Usp46
|
ubiquitin specific peptidase 46 |
| chr6_-_24563246 | 0.09 |
ENSRNOT00000074294
|
LOC685881
|
hypothetical protein LOC685881 |
| chr15_+_4209703 | 0.09 |
ENSRNOT00000082236
|
Ppp3cb
|
protein phosphatase 3 catalytic subunit beta |
| chrX_-_123254557 | 0.08 |
ENSRNOT00000039809
|
Akap17b
|
A kinase (PRKA) anchor protein 17B |
| chr5_-_79222687 | 0.08 |
ENSRNOT00000010516
|
Akna
|
AT-hook transcription factor |
| chr8_-_48597867 | 0.08 |
ENSRNOT00000077958
|
Nlrx1
|
NLR family member X1 |
| chrX_-_152642531 | 0.08 |
ENSRNOT00000085037
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr3_-_9326993 | 0.08 |
ENSRNOT00000090137
|
Lamc3
|
laminin subunit gamma 3 |
| chr9_-_27761733 | 0.08 |
ENSRNOT00000040034
|
Kcnq5
|
potassium voltage-gated channel subfamily Q member 5 |
| chrX_-_75566481 | 0.08 |
ENSRNOT00000003714
|
Zdhhc15
|
zinc finger, DHHC-type containing 15 |
| chr16_-_21362955 | 0.08 |
ENSRNOT00000039607
|
Gmip
|
Gem-interacting protein |
| chr19_-_37970537 | 0.08 |
ENSRNOT00000034722
|
Dpep2
|
dipeptidase 2 |
| chr4_-_168297373 | 0.08 |
ENSRNOT00000066575
|
Lrp6
|
LDL receptor related protein 6 |
| chr3_+_2182957 | 0.08 |
ENSRNOT00000011633
|
Pnpla7
|
patatin-like phospholipase domain containing 7 |
| chr9_-_81864202 | 0.08 |
ENSRNOT00000084964
|
Zfp142
|
zinc finger protein 142 |
| chr6_+_137184820 | 0.08 |
ENSRNOT00000073796
|
Adssl1
|
adenylosuccinate synthase like 1 |
| chr3_+_4106477 | 0.08 |
ENSRNOT00000078985
|
AABR07051251.1
|
|
| chr5_-_100647298 | 0.08 |
ENSRNOT00000067538
ENSRNOT00000013092 |
Nfib
|
nuclear factor I/B |
| chr1_+_114258719 | 0.08 |
ENSRNOT00000088459
ENSRNOT00000016376 |
Cyfip1
|
cytoplasmic FMR1 interacting protein 1 |
| chr13_+_90301006 | 0.08 |
ENSRNOT00000029315
|
Slamf6
|
SLAM family member 6 |
| chr2_+_211546560 | 0.08 |
ENSRNOT00000033443
|
Aknad1
|
AKNA domain containing 1 |
| chr7_-_62972084 | 0.08 |
ENSRNOT00000064251
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr3_+_72329967 | 0.07 |
ENSRNOT00000090256
|
Slc43a3
|
solute carrier family 43, member 3 |
| chr20_-_14025067 | 0.07 |
ENSRNOT00000093430
ENSRNOT00000074533 |
Ggt1
|
gamma-glutamyltransferase 1 |
| chr10_-_90501819 | 0.07 |
ENSRNOT00000050474
|
Gpatch8
|
G patch domain containing 8 |
| chr8_+_99880073 | 0.07 |
ENSRNOT00000010765
|
Plscr4
|
phospholipid scramblase 4 |
| chr3_-_97993684 | 0.07 |
ENSRNOT00000006481
|
Arl14ep
|
ADP-ribosylation factor like GTPase 14 effector protein |
| chr9_+_73529612 | 0.07 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr3_+_175450979 | 0.07 |
ENSRNOT00000089716
|
Mtg2
|
mitochondrial ribosome-associated GTPase 2 |
| chr8_-_105462141 | 0.07 |
ENSRNOT00000066731
ENSRNOT00000078760 |
Clstn2
|
calsyntenin 2 |
| chr8_-_76579387 | 0.07 |
ENSRNOT00000090747
|
Fam81a
|
family with sequence similarity 81, member A |
| chr15_-_8931983 | 0.07 |
ENSRNOT00000085322
|
Thrb
|
thyroid hormone receptor beta |
| chr1_+_5366379 | 0.07 |
ENSRNOT00000019918
|
Fbxo30
|
F-box protein 30 |
| chrX_+_22313028 | 0.07 |
ENSRNOT00000082725
|
Kdm5c
|
lysine demethylase 5C |
| chr19_-_19315357 | 0.07 |
ENSRNOT00000018888
|
Cyld
|
CYLD lysine 63 deubiquitinase |
| chrX_-_1786978 | 0.07 |
ENSRNOT00000011317
|
Rbm10
|
RNA binding motif protein 10 |
| chr2_-_88414012 | 0.07 |
ENSRNOT00000014762
|
Lrrcc1
|
leucine rich repeat and coiled-coil centrosomal protein 1 |
| chr3_+_72080630 | 0.07 |
ENSRNOT00000008911
|
Med19
|
mediator complex subunit 19 |
| chr8_-_104542317 | 0.07 |
ENSRNOT00000016327
|
Rasa2
|
RAS p21 protein activator 2 |
| chr13_+_89586283 | 0.07 |
ENSRNOT00000079355
ENSRNOT00000049873 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr5_-_160403373 | 0.07 |
ENSRNOT00000018393
|
Ctrc
|
chymotrypsin C |
| chr3_-_15278645 | 0.06 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr10_-_68926264 | 0.06 |
ENSRNOT00000008661
|
Phb-ps1
|
prohibitin, pseudogene 1 |
| chr2_-_22744407 | 0.06 |
ENSRNOT00000073710
|
Cmya5
|
cardiomyopathy associated 5 |
| chr2_-_33025271 | 0.06 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr8_-_76579099 | 0.06 |
ENSRNOT00000088628
|
Fam81a
|
family with sequence similarity 81, member A |
| chr8_+_107875991 | 0.06 |
ENSRNOT00000090299
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr19_-_50220455 | 0.06 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr6_+_145546595 | 0.06 |
ENSRNOT00000007112
|
Rapgef5
|
Rap guanine nucleotide exchange factor 5 |
| chr8_+_45797315 | 0.06 |
ENSRNOT00000059997
|
AABR07070046.1
|
|
| chr16_-_20460959 | 0.06 |
ENSRNOT00000026457
|
Pde4c
|
phosphodiesterase 4C |
| chr10_+_66942398 | 0.06 |
ENSRNOT00000018986
|
Rab11fip4
|
RAB11 family interacting protein 4 |
| chr15_-_52344629 | 0.06 |
ENSRNOT00000032832
|
Npm2
|
nucleophosmin/nucleoplasmin 2 |
| chr2_-_185303610 | 0.06 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr4_+_81311490 | 0.06 |
ENSRNOT00000016265
|
Snx10
|
sorting nexin 10 |
| chr6_+_26642783 | 0.06 |
ENSRNOT00000008126
|
Slc30a3
|
solute carrier family 30 member 3 |
| chr9_+_50664150 | 0.06 |
ENSRNOT00000015393
|
Tpp2
|
tripeptidyl peptidase 2 |
| chr6_-_122572925 | 0.06 |
ENSRNOT00000090869
|
AC123293.2
|
|
| chr6_-_114488880 | 0.06 |
ENSRNOT00000087560
|
AC118957.1
|
|
| chr3_+_8534440 | 0.06 |
ENSRNOT00000045827
ENSRNOT00000082672 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr1_+_81230989 | 0.05 |
ENSRNOT00000077952
|
Kcnn4
|
potassium calcium-activated channel subfamily N member 4 |
| chr4_-_6062641 | 0.05 |
ENSRNOT00000074846
|
Cct8l1
|
chaperonin containing TCP1, subunit 8 (theta)-like 1 |
| chr9_+_77834091 | 0.05 |
ENSRNOT00000033459
|
Vwc2l
|
von Willebrand factor C domain-containing protein 2-like |
| chr11_-_17130120 | 0.05 |
ENSRNOT00000002119
|
RGD1563888
|
similar to DNA segment, Chr 16, ERATO Doi 472, expressed |
| chr10_+_108132105 | 0.05 |
ENSRNOT00000072534
|
Cbx2
|
chromobox 2 |
| chr1_+_154377447 | 0.05 |
ENSRNOT00000084268
ENSRNOT00000092086 ENSRNOT00000091470 ENSRNOT00000025415 |
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr11_-_39767802 | 0.05 |
ENSRNOT00000072806
|
RGD1565472
|
similar to RIKEN cDNA 2310003P10 |
| chr7_-_116872511 | 0.05 |
ENSRNOT00000010120
|
Zc3h3
|
zinc finger CCCH type containing 3 |
| chr7_-_140454268 | 0.05 |
ENSRNOT00000081468
|
Wnt10b
|
wingless-type MMTV integration site family, member 10B |
| chr16_+_80729400 | 0.05 |
ENSRNOT00000036383
|
Tdrp
|
testis development related protein |
| chr4_-_33761163 | 0.05 |
ENSRNOT00000010079
|
Asns
|
asparagine synthetase (glutamine-hydrolyzing) |
| chr12_+_37594185 | 0.05 |
ENSRNOT00000088787
|
Sbno1
|
strawberry notch homolog 1 |
| chr4_+_41364441 | 0.05 |
ENSRNOT00000087146
|
Foxp2
|
forkhead box P2 |
| chr2_-_189096785 | 0.05 |
ENSRNOT00000028200
|
Chrnb2
|
cholinergic receptor nicotinic beta 2 subunit |
| chr5_+_35916804 | 0.05 |
ENSRNOT00000066043
ENSRNOT00000083132 |
Usp45
|
ubiquitin specific peptidase 45 |
| chr11_-_72378718 | 0.05 |
ENSRNOT00000078737
|
Dlg1
|
discs large MAGUK scaffold protein 1 |
| chr20_-_4542073 | 0.05 |
ENSRNOT00000000477
|
Cfb
|
complement factor B |
| chr5_+_146383942 | 0.05 |
ENSRNOT00000078588
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr8_+_79323408 | 0.05 |
ENSRNOT00000088391
ENSRNOT00000090258 |
Nedd4
|
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr4_-_78924181 | 0.05 |
ENSRNOT00000012325
|
Tra2a
|
transformer 2 alpha homolog |
| chr13_+_89597138 | 0.05 |
ENSRNOT00000004662
|
Apoa2
|
apolipoprotein A2 |
| chr4_-_51943519 | 0.05 |
ENSRNOT00000060459
|
Pot1
|
protection of telomeres 1 |
| chr18_-_70924708 | 0.05 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr2_-_23260965 | 0.05 |
ENSRNOT00000046772
|
LOC103691423
|
60S ribosomal protein L36 pseudogene |
| chr1_+_78767911 | 0.05 |
ENSRNOT00000022360
|
Prkd2
|
protein kinase D2 |
| chr1_+_167522895 | 0.05 |
ENSRNOT00000092987
|
Stim1
|
stromal interaction molecule 1 |
| chr1_+_215609645 | 0.05 |
ENSRNOT00000076140
ENSRNOT00000027487 |
Tnni2
|
troponin I2, fast skeletal type |
| chr4_-_157155609 | 0.04 |
ENSRNOT00000016330
|
C1s
|
complement C1s |
| chr18_+_27576129 | 0.04 |
ENSRNOT00000070930
|
Kdm3b
|
lysine demethylase 3B |
| chrX_+_128416722 | 0.04 |
ENSRNOT00000009336
ENSRNOT00000085110 |
Xiap
|
X-linked inhibitor of apoptosis |
| chr6_+_47916188 | 0.04 |
ENSRNOT00000011819
|
Rnaseh1
|
ribonuclease H1 |
| chr1_+_199225100 | 0.04 |
ENSRNOT00000088606
|
Setd1a
|
SET domain containing 1A |
| chr2_+_18354542 | 0.04 |
ENSRNOT00000042958
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr20_-_5927070 | 0.04 |
ENSRNOT00000059264
|
Slc26a8
|
solute carrier family 26 member 8 |
| chr1_-_42587666 | 0.04 |
ENSRNOT00000083225
ENSRNOT00000025355 |
Rgs17
|
regulator of G-protein signaling 17 |
| chr10_+_86657285 | 0.04 |
ENSRNOT00000087346
|
Thra
|
thyroid hormone receptor alpha |
| chr20_+_4363152 | 0.04 |
ENSRNOT00000000508
ENSRNOT00000084841 ENSRNOT00000072848 ENSRNOT00000077561 |
Ager
|
advanced glycosylation end product-specific receptor |
| chr6_+_44225233 | 0.04 |
ENSRNOT00000066593
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr1_-_88361712 | 0.04 |
ENSRNOT00000033661
ENSRNOT00000083788 |
Zfp82
|
zinc finger protein 82 |
| chr2_-_27296777 | 0.04 |
ENSRNOT00000080748
|
Ankdd1b
|
ankyrin repeat and death domain containing 1B |
| chr9_-_116222374 | 0.04 |
ENSRNOT00000090111
ENSRNOT00000067900 |
Arhgap28
|
Rho GTPase activating protein 28 |
| chr1_+_198655742 | 0.04 |
ENSRNOT00000023944
|
Mylpf
|
myosin light chain, phosphorylatable, fast skeletal muscle |
| chr2_+_198721724 | 0.04 |
ENSRNOT00000043535
|
Ankrd34a
|
ankyrin repeat domain 34A |
| chr10_+_20320878 | 0.04 |
ENSRNOT00000009714
|
Slit3
|
slit guidance ligand 3 |
| chr11_+_89277039 | 0.04 |
ENSRNOT00000002511
|
Mzt2b
|
mitotic spindle organizing protein 2B |
| chr1_+_256382791 | 0.04 |
ENSRNOT00000022549
|
Cyp26a1
|
cytochrome P450, family 26, subfamily a, polypeptide 1 |
| chr17_-_42276797 | 0.04 |
ENSRNOT00000083996
|
LOC100911664
|
uncharacterized LOC100911664 |
| chr16_-_7290561 | 0.04 |
ENSRNOT00000036910
|
Nisch
|
nischarin |
| chr3_+_95959703 | 0.04 |
ENSRNOT00000006406
|
Immp1l
|
inner mitochondrial membrane peptidase subunit 1 |
| chr2_-_189899325 | 0.04 |
ENSRNOT00000017561
|
Chtop
|
chromatin target of PRMT1 |
| chr7_-_70661891 | 0.04 |
ENSRNOT00000010240
|
Inhbc
|
inhibin beta C subunit |
| chr2_+_49682754 | 0.04 |
ENSRNOT00000079907
ENSRNOT00000085576 |
Emb
|
embigin |
| chr7_-_143538579 | 0.03 |
ENSRNOT00000081518
|
Krt79
|
keratin 79 |
| chr8_+_39878955 | 0.03 |
ENSRNOT00000060300
|
LOC100911068
|
roundabout homolog 4-like |
| chr1_-_143315633 | 0.03 |
ENSRNOT00000026030
|
Ap3b2
|
adaptor-related protein complex 3, beta 2 subunit |
| chr8_-_22259779 | 0.03 |
ENSRNOT00000028360
|
Keap1
|
Kelch-like ECH-associated protein 1 |
| chr5_-_57683932 | 0.03 |
ENSRNOT00000074796
|
Dcaf12
|
DDB1 and CUL4 associated factor 12 |
| chr8_+_63036976 | 0.03 |
ENSRNOT00000011383
|
Stoml1
|
stomatin like 1 |
| chr7_-_116504853 | 0.03 |
ENSRNOT00000056557
|
RGD1565410
|
similar to Ly6-C antigen gene |
| chr2_+_62150493 | 0.03 |
ENSRNOT00000080241
|
Zfr
|
zinc finger RNA binding protein |
| chr1_+_215609036 | 0.03 |
ENSRNOT00000076187
|
Tnni2
|
troponin I2, fast skeletal type |
| chr4_-_157433467 | 0.03 |
ENSRNOT00000028965
|
Lag3
|
lymphocyte activating 3 |
| chr18_+_59748444 | 0.03 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr11_-_84068302 | 0.03 |
ENSRNOT00000002323
|
Dvl3
|
dishevelled segment polarity protein 3 |
| chr1_-_127599257 | 0.03 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr8_-_128622314 | 0.03 |
ENSRNOT00000084950
ENSRNOT00000024408 |
Gorasp1
|
golgi reassembly stacking protein 1 |
| chr19_-_32528965 | 0.03 |
ENSRNOT00000015929
|
Zfp827
|
zinc finger protein 827 |
| chr1_-_131460473 | 0.03 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr8_+_130401470 | 0.03 |
ENSRNOT00000043346
|
Zbtb47
|
zinc finger and BTB domain containing 47 |
| chr17_+_47241017 | 0.03 |
ENSRNOT00000092193
|
Gpr141
|
G protein-coupled receptor 141 |
| chr17_+_49322205 | 0.03 |
ENSRNOT00000017713
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr4_-_146839397 | 0.03 |
ENSRNOT00000010338
|
Vgll4
|
vestigial-like family member 4 |
| chr10_-_70341837 | 0.03 |
ENSRNOT00000077261
|
Slfn13
|
schlafen family member 13 |
| chr13_+_40300039 | 0.03 |
ENSRNOT00000049137
|
AABR07020786.1
|
|
| chr12_-_1816414 | 0.03 |
ENSRNOT00000041155
ENSRNOT00000067448 |
Insr
|
insulin receptor |
| chr10_+_83231238 | 0.03 |
ENSRNOT00000077625
|
Spop
|
speckle type BTB/POZ protein |
| chr3_+_35679750 | 0.03 |
ENSRNOT00000059588
|
Lypd6
|
LY6/PLAUR domain containing 6 |
| chrX_+_71528988 | 0.03 |
ENSRNOT00000075963
|
Ogt
|
O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr14_-_45859908 | 0.03 |
ENSRNOT00000086994
|
Pgm2
|
phosphoglucomutase 2 |
| chr14_+_84447885 | 0.03 |
ENSRNOT00000009150
|
Gatsl3
|
GATS protein-like 3 |
| chr5_-_158313426 | 0.03 |
ENSRNOT00000025488
|
Pax7
|
paired box 7 |
| chr7_-_12634641 | 0.03 |
ENSRNOT00000093132
|
Cfd
|
complement factor D |
| chr8_-_114013623 | 0.03 |
ENSRNOT00000018175
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr10_-_15211325 | 0.03 |
ENSRNOT00000027083
|
Rhot2
|
ras homolog family member T2 |
| chr16_+_8497569 | 0.03 |
ENSRNOT00000027054
|
Ogdhl
|
oxoglutarate dehydrogenase-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rreb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0021740 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.1 | 0.2 | GO:0019389 | glucuronoside metabolic process(GO:0019389) response to 2,3,7,8-tetrachlorodibenzodioxine(GO:1904612) |
| 0.1 | 0.2 | GO:1905204 | response to selenite ion(GO:0072714) negative regulation of connective tissue replacement(GO:1905204) |
| 0.1 | 0.8 | GO:0043129 | surfactant homeostasis(GO:0043129) |
| 0.0 | 0.2 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 0.1 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 0.0 | 0.1 | GO:1903116 | positive regulation of actin filament-based movement(GO:1903116) |
| 0.0 | 0.2 | GO:0033668 | negative regulation by symbiont of host apoptotic process(GO:0033668) negative regulation by symbiont of host programmed cell death(GO:0052041) negative regulation by organism of programmed cell death in other organism involved in symbiotic interaction(GO:0052490) |
| 0.0 | 0.2 | GO:0048669 | somite specification(GO:0001757) collateral sprouting in absence of injury(GO:0048669) |
| 0.0 | 0.1 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.1 | GO:1903756 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0035279 | mRNA cleavage involved in gene silencing by miRNA(GO:0035279) mRNA cleavage involved in gene silencing(GO:0098795) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.1 | GO:1901219 | external genitalia morphogenesis(GO:0035261) canonical Wnt signaling pathway involved in positive regulation of cardiac outflow tract cell proliferation(GO:0061324) regulation of cardiac chamber morphogenesis(GO:1901219) negative regulation of cardiac chamber morphogenesis(GO:1901220) regulation of cardiac ventricle development(GO:1904412) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:0044208 | 'de novo' AMP biosynthetic process(GO:0044208) |
| 0.0 | 0.1 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) |
| 0.0 | 0.1 | GO:1903760 | regulation of voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1903760) |
| 0.0 | 0.1 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.0 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0002540 | leukotriene production involved in inflammatory response(GO:0002540) |
| 0.0 | 0.2 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:1902963 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.1 | GO:0031179 | peptide modification(GO:0031179) |
| 0.0 | 0.1 | GO:2001184 | positive regulation of interleukin-12 secretion(GO:2001184) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0006529 | asparagine biosynthetic process(GO:0006529) |
| 0.0 | 0.1 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.0 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 0.0 | GO:0060383 | positive regulation of DNA strand elongation(GO:0060383) |
| 0.0 | 0.1 | GO:0043102 | amino acid salvage(GO:0043102) L-methionine salvage(GO:0071267) |
| 0.0 | 0.1 | GO:1990785 | response to water-immersion restraint stress(GO:1990785) |
| 0.0 | 0.2 | GO:0006688 | glycosphingolipid biosynthetic process(GO:0006688) |
| 0.0 | 0.1 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.0 | GO:0038033 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) |
| 0.0 | 0.1 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.0 | 0.0 | GO:0043137 | DNA replication, removal of RNA primer(GO:0043137) |
| 0.0 | 0.1 | GO:0070885 | negative regulation of calcineurin-NFAT signaling cascade(GO:0070885) |
| 0.0 | 0.0 | GO:0060849 | lymphatic endothelial cell fate commitment(GO:0060838) regulation of transcription involved in lymphatic endothelial cell fate commitment(GO:0060849) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:0097208 | alveolar lamellar body(GO:0097208) |
| 0.0 | 0.3 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.2 | GO:0050785 | advanced glycation end-product receptor activity(GO:0050785) |
| 0.0 | 0.1 | GO:0008489 | UDP-galactose:glucosylceramide beta-1,4-galactosyltransferase activity(GO:0008489) |
| 0.0 | 0.8 | GO:0004190 | aspartic-type endopeptidase activity(GO:0004190) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0005030 | neurotrophin receptor activity(GO:0005030) |
| 0.0 | 0.1 | GO:0004051 | arachidonate 5-lipoxygenase activity(GO:0004051) |
| 0.0 | 0.2 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.1 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.0 | 0.1 | GO:0004066 | asparagine synthase (glutamine-hydrolyzing) activity(GO:0004066) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.2 | GO:0004887 | thyroid hormone receptor activity(GO:0004887) |
| 0.0 | 0.1 | GO:0008240 | tripeptidyl-peptidase activity(GO:0008240) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.0 | GO:1990955 | G-rich single-stranded DNA binding(GO:1990955) |
| 0.0 | 0.0 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.0 | 0.1 | GO:0003840 | gamma-glutamyltransferase activity(GO:0003840) glutathione hydrolase activity(GO:0036374) |
| 0.0 | 0.2 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.3 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 0.2 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.2 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |