Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Rorc_Nr1d1
Z-value: 0.39
Transcription factors associated with Rorc_Nr1d1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
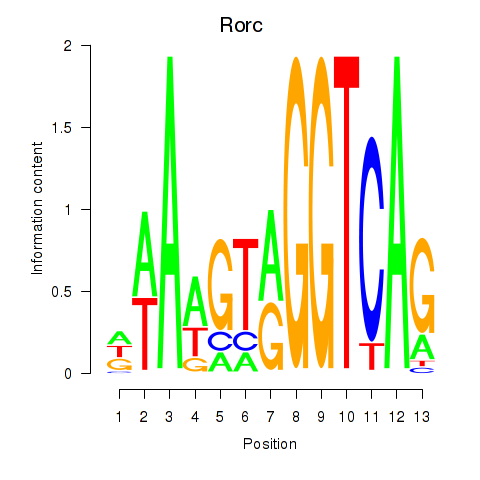
Rorc
|
ENSRNOG00000020836 | RAR-related orphan receptor C |
|
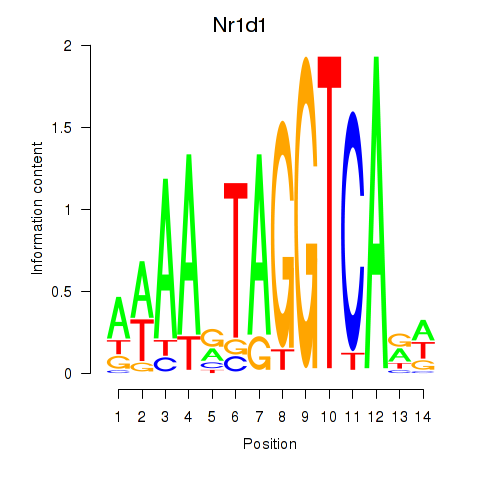
Nr1d1
|
ENSRNOG00000009329 | nuclear receptor subfamily 1, group D, member 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rorc | rn6_v1_chr2_+_195617021_195617044 | 0.24 | 7.0e-01 | Click! |
| Nr1d1 | rn6_v1_chr10_-_86690815_86690815 | -0.11 | 8.6e-01 | Click! |
Activity profile of Rorc_Nr1d1 motif
Sorted Z-values of Rorc_Nr1d1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_79713567 | 0.41 |
ENSRNOT00000012110
|
C1qtnf4
|
C1q and tumor necrosis factor related protein 4 |
| chr12_-_2174131 | 0.23 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr1_-_72339395 | 0.17 |
ENSRNOT00000021772
|
Zfp580
|
zinc finger protein 580 |
| chr19_-_25062934 | 0.11 |
ENSRNOT00000060117
|
Gm10644
|
predicted gene 10644 |
| chr10_+_14248399 | 0.11 |
ENSRNOT00000077689
|
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr3_+_2642531 | 0.11 |
ENSRNOT00000081798
|
Fut7
|
fucosyltransferase 7 |
| chr5_-_72287669 | 0.11 |
ENSRNOT00000022255
|
Klf4
|
Kruppel like factor 4 |
| chr20_+_44680449 | 0.10 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr1_-_225594958 | 0.06 |
ENSRNOT00000027492
|
Scgb1d2
|
secretoglobin, family 1D, member 2 |
| chr11_+_87366621 | 0.06 |
ENSRNOT00000040978
|
Aifm3
|
apoptosis inducing factor, mitochondria associated 3 |
| chr4_+_199916 | 0.06 |
ENSRNOT00000009317
|
Htr5a
|
5-hydroxytryptamine receptor 5A |
| chr8_-_109418872 | 0.06 |
ENSRNOT00000021657
|
Pccb
|
propionyl-CoA carboxylase beta subunit |
| chr9_+_17340341 | 0.06 |
ENSRNOT00000026637
ENSRNOT00000026559 ENSRNOT00000042790 ENSRNOT00000044163 ENSRNOT00000083811 |
Vegfa
|
vascular endothelial growth factor A |
| chr2_+_122877286 | 0.05 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr18_-_55771730 | 0.05 |
ENSRNOT00000026426
|
Smim3
|
small integral membrane protein 3 |
| chr8_+_48443767 | 0.05 |
ENSRNOT00000010127
|
C1qtnf5
|
C1q and tumor necrosis factor related protein 5 |
| chr2_-_260115577 | 0.05 |
ENSRNOT00000065997
|
Rabggtb
|
Rab geranylgeranyltransferase, beta subunit |
| chr8_+_82380757 | 0.05 |
ENSRNOT00000013512
|
Leo1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr2_-_189096785 | 0.05 |
ENSRNOT00000028200
|
Chrnb2
|
cholinergic receptor nicotinic beta 2 subunit |
| chr4_-_123510217 | 0.05 |
ENSRNOT00000080734
|
Slc41a3
|
solute carrier family 41, member 3 |
| chr19_-_26194198 | 0.05 |
ENSRNOT00000005917
|
Wdr83
|
WD repeat domain 83 |
| chr18_+_30527705 | 0.05 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr6_+_104437159 | 0.04 |
ENSRNOT00000076485
|
Susd6
|
sushi domain containing 6 |
| chr1_-_88066101 | 0.04 |
ENSRNOT00000079473
ENSRNOT00000027893 |
Ryr1
|
ryanodine receptor 1 |
| chr2_+_189997129 | 0.03 |
ENSRNOT00000015958
|
S100a4
|
S100 calcium-binding protein A4 |
| chrX_+_33895769 | 0.03 |
ENSRNOT00000085993
ENSRNOT00000079365 |
Reps2
|
RALBP1 associated Eps domain containing protein 2 |
| chr2_+_252263386 | 0.03 |
ENSRNOT00000092913
ENSRNOT00000084034 ENSRNOT00000041186 ENSRNOT00000092931 |
Ssx2ip
|
SSX family member 2 interacting protein |
| chr13_-_49169918 | 0.03 |
ENSRNOT00000000036
|
Tmcc2
|
transmembrane and coiled-coil domain family 2 |
| chr1_-_166037424 | 0.03 |
ENSRNOT00000026115
|
P2ry2
|
purinergic receptor P2Y2 |
| chrX_+_136488691 | 0.03 |
ENSRNOT00000093432
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr14_-_72970329 | 0.03 |
ENSRNOT00000006325
|
Pp2d1
|
protein phosphatase 2C-like domain containing 1 |
| chr7_-_101140308 | 0.03 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chrX_+_106791333 | 0.03 |
ENSRNOT00000050302
|
Tceal9
|
transcription elongation factor A like 9 |
| chr10_+_14248710 | 0.02 |
ENSRNOT00000020733
|
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr19_-_37725623 | 0.02 |
ENSRNOT00000024197
|
Gfod2
|
glucose-fructose oxidoreductase domain containing 2 |
| chr1_-_214414763 | 0.02 |
ENSRNOT00000025240
ENSRNOT00000077776 |
LOC100911440
|
mitochondrial glutamate carrier 1-like |
| chr1_+_185356975 | 0.02 |
ENSRNOT00000086681
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr11_-_39767802 | 0.02 |
ENSRNOT00000072806
|
RGD1565472
|
similar to RIKEN cDNA 2310003P10 |
| chr1_+_87066289 | 0.02 |
ENSRNOT00000027645
|
Capn12
|
calpain 12 |
| chr4_+_24612205 | 0.02 |
ENSRNOT00000057382
|
Ewsr1
|
EWS RNA-binding protein 1 |
| chr1_-_91588609 | 0.02 |
ENSRNOT00000050931
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr2_+_60461551 | 0.02 |
ENSRNOT00000024557
|
Rad1
|
RAD1 checkpoint DNA exonuclease |
| chr1_-_67065797 | 0.02 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr9_-_88534710 | 0.02 |
ENSRNOT00000020880
|
Tm4sf20
|
transmembrane 4 L six family member 20 |
| chr10_-_94365049 | 0.02 |
ENSRNOT00000011894
|
Strada
|
STE20-related kinase adaptor alpha |
| chr1_+_53174879 | 0.02 |
ENSRNOT00000017601
ENSRNOT00000084628 |
Rnaset2
|
ribonuclease T2 |
| chr8_+_48569328 | 0.02 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr1_+_102017263 | 0.02 |
ENSRNOT00000028680
|
Nomo1
|
nodal modulator 1 |
| chr7_+_64672722 | 0.02 |
ENSRNOT00000064448
ENSRNOT00000005539 |
Grip1
|
glutamate receptor interacting protein 1 |
| chr10_-_5511955 | 0.01 |
ENSRNOT00000039429
|
Atf7ip2
|
activating transcription factor 7 interacting protein 2 |
| chr1_-_20962526 | 0.01 |
ENSRNOT00000061332
ENSRNOT00000017322 ENSRNOT00000017412 ENSRNOT00000079688 ENSRNOT00000017417 |
Epb41l2
|
erythrocyte membrane protein band 4.1-like 2 |
| chr4_-_117751329 | 0.01 |
ENSRNOT00000080096
|
LOC100910829
|
probable N-acetyltransferase CML2-like |
| chr1_+_102857251 | 0.01 |
ENSRNOT00000087920
|
Gtf2h1
|
general transcription factor IIH subunit 1 |
| chr7_-_114573900 | 0.01 |
ENSRNOT00000011219
|
Ptk2
|
protein tyrosine kinase 2 |
| chrX_+_40286592 | 0.01 |
ENSRNOT00000087898
|
Mbtps2
|
membrane-bound transcription factor peptidase, site 2 |
| chr2_-_157759819 | 0.01 |
ENSRNOT00000015763
ENSRNOT00000016016 |
LOC100909712
|
cyclin-L1-like |
| chr3_+_59408546 | 0.01 |
ENSRNOT00000031418
|
AABR07052523.1
|
|
| chr1_-_265542276 | 0.01 |
ENSRNOT00000065172
|
Mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chr3_+_56056925 | 0.01 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr10_-_18131562 | 0.01 |
ENSRNOT00000006676
|
Tlx3
|
T-cell leukemia, homeobox 3 |
| chr3_+_139894331 | 0.01 |
ENSRNOT00000064695
|
Rin2
|
Ras and Rab interactor 2 |
| chr7_-_12405022 | 0.01 |
ENSRNOT00000021648
|
Cirbp
|
cold inducible RNA binding protein |
| chr10_-_88442796 | 0.01 |
ENSRNOT00000077579
ENSRNOT00000023447 ENSRNOT00000043172 |
Acly
|
ATP citrate lyase |
| chrX_-_72133692 | 0.01 |
ENSRNOT00000004263
|
Cited1
|
Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 |
| chr5_-_88612626 | 0.01 |
ENSRNOT00000089560
|
Tle1
|
transducin like enhancer of split 1 |
| chr9_-_40008680 | 0.01 |
ENSRNOT00000016578
|
Khdrbs2
|
KH RNA binding domain containing, signal transduction associated 2 |
| chr17_-_12669573 | 0.01 |
ENSRNOT00000016942
ENSRNOT00000041726 |
Syk
|
spleen associated tyrosine kinase |
| chr4_-_124048417 | 0.01 |
ENSRNOT00000014933
|
Mrps25
|
mitochondrial ribosomal protein S25 |
| chr10_+_86303727 | 0.01 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr14_+_46649971 | 0.01 |
ENSRNOT00000085875
|
AABR07015078.1
|
|
| chr7_-_136853154 | 0.01 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr8_+_127171537 | 0.01 |
ENSRNOT00000078625
ENSRNOT00000051290 ENSRNOT00000087753 |
Golga4
|
golgin A4 |
| chr14_+_83889089 | 0.01 |
ENSRNOT00000078980
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr14_-_21299068 | 0.00 |
ENSRNOT00000065778
|
Amtn
|
amelotin |
| chr4_+_44774741 | 0.00 |
ENSRNOT00000086902
|
Met
|
MET proto-oncogene, receptor tyrosine kinase |
| chr6_-_146470456 | 0.00 |
ENSRNOT00000018479
|
RGD1560883
|
similar to KIAA0825 protein |
| chr1_-_90520344 | 0.00 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr1_-_90520012 | 0.00 |
ENSRNOT00000028698
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr1_-_274106502 | 0.00 |
ENSRNOT00000020539
|
Smndc1
|
survival motor neuron domain containing 1 |
| chr18_-_79258570 | 0.00 |
ENSRNOT00000022401
|
Galr1
|
galanin receptor 1 |
| chr1_-_170404056 | 0.00 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr2_+_178679041 | 0.00 |
ENSRNOT00000013524
|
Fam198b
|
family with sequence similarity 198, member B |
| chr14_+_100311173 | 0.00 |
ENSRNOT00000031275
|
Ppp3r1
|
protein phosphatase 3, regulatory subunit B, alpha |
| chr18_-_86279680 | 0.00 |
ENSRNOT00000006169
|
LOC689166
|
hypothetical protein LOC689166 |
| chr14_-_84334066 | 0.00 |
ENSRNOT00000006160
|
Mtfp1
|
mitochondrial fission process 1 |
| chr5_-_50684409 | 0.00 |
ENSRNOT00000013181
|
RGD1359108
|
similar to RIKEN cDNA 3110043O21 |
| chr2_+_142686724 | 0.00 |
ENSRNOT00000014614
|
Proser1
|
proline and serine rich 1 |
| chr12_-_46809849 | 0.00 |
ENSRNOT00000081134
|
Pxn
|
paxillin |
| chr7_-_18585313 | 0.00 |
ENSRNOT00000010399
|
March2
|
membrane associated ring-CH-type finger 2 |
| chr9_+_65172194 | 0.00 |
ENSRNOT00000040493
|
AC128084.1
|
|
| chr4_-_125929002 | 0.00 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr1_+_250426158 | 0.00 |
ENSRNOT00000067643
|
A1cf
|
APOBEC1 complementation factor |
| chr2_+_61991742 | 0.00 |
ENSRNOT00000046715
|
LOC499544
|
LRRGT00154 |
| chrX_-_1952111 | 0.00 |
ENSRNOT00000061140
|
Jade3
|
jade family PHD finger 3 |
| chrX_+_6791136 | 0.00 |
ENSRNOT00000003984
|
LOC100909913
|
norrin-like |
| chr8_-_68275720 | 0.00 |
ENSRNOT00000079122
|
Map2k5
|
mitogen activated protein kinase kinase 5 |
| chr10_-_72417070 | 0.00 |
ENSRNOT00000036814
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr4_-_157230647 | 0.00 |
ENSRNOT00000017334
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rorc_Nr1d1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:1901492 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) cardiac vascular smooth muscle cell development(GO:0060948) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.0 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.0 | 0.1 | GO:0007198 | adenylate cyclase-inhibiting serotonin receptor signaling pathway(GO:0007198) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.0 | 0.0 | GO:1990269 | RNA polymerase II C-terminal domain phosphoserine binding(GO:1990269) |