Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
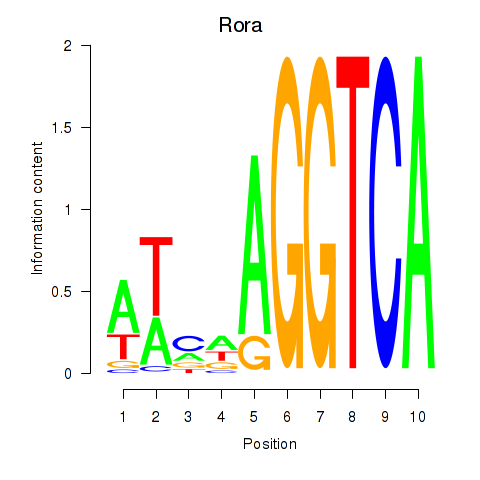
Results for Rora
Z-value: 0.34
Transcription factors associated with Rora
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rora
|
ENSRNOG00000027145 | RAR-related orphan receptor A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rora | rn6_v1_chr8_+_75516904_75516904 | 0.05 | 9.4e-01 | Click! |
Activity profile of Rora motif
Sorted Z-values of Rora motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_+_61531416 | 0.15 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr4_+_51614676 | 0.14 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr8_+_29453643 | 0.12 |
ENSRNOT00000090643
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr10_-_85517683 | 0.11 |
ENSRNOT00000016070
|
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr3_-_93789617 | 0.11 |
ENSRNOT00000078616
|
Caprin1
|
cell cycle associated protein 1 |
| chr7_+_121311024 | 0.10 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr1_+_100593892 | 0.10 |
ENSRNOT00000027062
|
Kcnc3
|
potassium voltage-gated channel subfamily C member 3 |
| chr6_-_76552559 | 0.10 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr12_+_40018937 | 0.09 |
ENSRNOT00000001697
|
Cux2
|
cut-like homeobox 2 |
| chr3_+_134413170 | 0.09 |
ENSRNOT00000074338
|
AABR07054000.1
|
|
| chr1_+_100593680 | 0.09 |
ENSRNOT00000078153
ENSRNOT00000027063 |
Kcnc3
|
potassium voltage-gated channel subfamily C member 3 |
| chr3_-_80012750 | 0.08 |
ENSRNOT00000018154
|
Nr1h3
|
nuclear receptor subfamily 1, group H, member 3 |
| chr8_-_82533689 | 0.08 |
ENSRNOT00000014124
|
Tmod2
|
tropomodulin 2 |
| chr3_-_146396299 | 0.08 |
ENSRNOT00000040188
ENSRNOT00000008931 |
Apmap
|
adipocyte plasma membrane associated protein |
| chr3_+_177226417 | 0.08 |
ENSRNOT00000031328
|
Oprl1
|
opioid related nociceptin receptor 1 |
| chr1_-_49844547 | 0.08 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr16_-_32753278 | 0.08 |
ENSRNOT00000015759
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr6_-_3355339 | 0.08 |
ENSRNOT00000084602
|
Map4k3
|
mitogen-activated protein kinase kinase kinase kinase 3 |
| chr10_-_86688730 | 0.08 |
ENSRNOT00000055333
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr10_-_90240509 | 0.08 |
ENSRNOT00000028407
|
Atxn7l3
|
ataxin 7-like 3 |
| chr8_+_114867062 | 0.08 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr5_+_147069616 | 0.07 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr2_+_28460068 | 0.07 |
ENSRNOT00000066819
|
Foxd1
|
forkhead box D1 |
| chr7_-_81592206 | 0.07 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr2_-_113345577 | 0.07 |
ENSRNOT00000034096
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr14_-_114583122 | 0.07 |
ENSRNOT00000084595
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr15_+_61069581 | 0.07 |
ENSRNOT00000084333
|
Vwa8
|
von Willebrand factor A domain containing 8 |
| chr1_+_224882439 | 0.06 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr20_+_28572242 | 0.06 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr1_+_197659187 | 0.06 |
ENSRNOT00000082228
|
LOC103690016
|
serine/threonine-protein kinase SBK1 |
| chr10_-_104748003 | 0.06 |
ENSRNOT00000042372
ENSRNOT00000046754 |
Acox1
|
acyl-CoA oxidase 1 |
| chr3_+_14304591 | 0.06 |
ENSRNOT00000007776
|
Cntrl
|
centriolin |
| chr2_-_56679955 | 0.06 |
ENSRNOT00000016722
|
Egflam
|
EGF-like, fibronectin type III and laminin G domains |
| chr1_-_221041401 | 0.06 |
ENSRNOT00000064136
|
Pcnx3
|
pecanex homolog 3 (Drosophila) |
| chr3_-_72113680 | 0.06 |
ENSRNOT00000009708
|
Zdhhc5
|
zinc finger, DHHC-type containing 5 |
| chr10_+_55712043 | 0.06 |
ENSRNOT00000010141
|
Aloxe3
|
arachidonate lipoxygenase 3 |
| chr3_+_151126591 | 0.06 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr3_-_60765645 | 0.06 |
ENSRNOT00000050513
|
Atf2
|
activating transcription factor 2 |
| chr1_-_73732118 | 0.06 |
ENSRNOT00000077964
|
Leng8
|
leukocyte receptor cluster member 8 |
| chr7_-_62972084 | 0.05 |
ENSRNOT00000064251
|
Msrb3
|
methionine sulfoxide reductase B3 |
| chr1_-_88066101 | 0.05 |
ENSRNOT00000079473
ENSRNOT00000027893 |
Ryr1
|
ryanodine receptor 1 |
| chr18_+_30856847 | 0.05 |
ENSRNOT00000027019
|
AABR07031734.1
|
|
| chr1_+_205706468 | 0.05 |
ENSRNOT00000089957
ENSRNOT00000023877 |
Edrf1
|
erythroid differentiation regulatory factor 1 |
| chr5_-_147412705 | 0.05 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr7_-_12393266 | 0.05 |
ENSRNOT00000021709
|
Efna2
|
ephrin A2 |
| chr11_-_87977368 | 0.05 |
ENSRNOT00000002546
|
Tmem191c
|
transmembrane protein 191C |
| chr10_+_59000397 | 0.05 |
ENSRNOT00000021134
|
Mybbp1a
|
MYB binding protein 1a |
| chr17_-_18592750 | 0.05 |
ENSRNOT00000065742
|
Cap2
|
CAP, adenylate cyclase-associated protein, 2 (yeast) |
| chr7_-_12424367 | 0.05 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr13_-_86451002 | 0.05 |
ENSRNOT00000043004
ENSRNOT00000027996 |
Pbx1
|
PBX homeobox 1 |
| chr9_+_10471742 | 0.05 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chr7_-_114590119 | 0.05 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr6_+_12362813 | 0.05 |
ENSRNOT00000022370
|
Ston1
|
stonin 1 |
| chr4_+_49369296 | 0.05 |
ENSRNOT00000007822
|
Wnt16
|
wingless-type MMTV integration site family, member 16 |
| chr8_+_59900651 | 0.04 |
ENSRNOT00000020410
|
Tmem266
|
transmembrane protein 266 |
| chr8_+_33239139 | 0.04 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr8_+_117280705 | 0.04 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr3_+_155297566 | 0.04 |
ENSRNOT00000021435
ENSRNOT00000084866 |
Dhx35
|
DEAH-box helicase 35 |
| chr3_-_112789282 | 0.04 |
ENSRNOT00000090680
ENSRNOT00000066181 |
Ttbk2
|
tau tubulin kinase 2 |
| chr1_-_222350173 | 0.04 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr19_-_26053762 | 0.04 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr6_-_28994519 | 0.04 |
ENSRNOT00000006668
|
Ubxn2a
|
UBX domain protein 2A |
| chr7_+_73273985 | 0.03 |
ENSRNOT00000077730
|
Pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr10_-_82399484 | 0.03 |
ENSRNOT00000082972
ENSRNOT00000055582 |
Xylt2
|
xylosyltransferase 2 |
| chr20_+_3351303 | 0.03 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr3_-_113525864 | 0.03 |
ENSRNOT00000046659
|
Frmd5
|
FERM domain containing 5 |
| chr1_-_78968329 | 0.03 |
ENSRNOT00000023078
|
Ppp5c
|
protein phosphatase 5, catalytic subunit |
| chr14_+_4125380 | 0.03 |
ENSRNOT00000002882
|
Zfp644
|
zinc finger protein 644 |
| chr1_-_265542276 | 0.03 |
ENSRNOT00000065172
|
Mgea5
|
meningioma expressed antigen 5 (hyaluronidase) |
| chrX_+_25016401 | 0.03 |
ENSRNOT00000059270
|
Clcn4
|
chloride voltage-gated channel 4 |
| chr13_+_110677810 | 0.03 |
ENSRNOT00000006340
|
Slc30a1
|
solute carrier family 30 member 1 |
| chr6_+_106496992 | 0.03 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr5_-_147148291 | 0.03 |
ENSRNOT00000085151
|
Azin2
|
antizyme inhibitor 2 |
| chr3_+_156727642 | 0.03 |
ENSRNOT00000078909
|
Plcg1
|
phospholipase C, gamma 1 |
| chr10_-_45579029 | 0.03 |
ENSRNOT00000080028
|
Arf1
|
ADP-ribosylation factor 1 |
| chr9_+_40975836 | 0.03 |
ENSRNOT00000084470
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr1_+_198682230 | 0.03 |
ENSRNOT00000023995
|
Znf48
|
zinc finger protein 48 |
| chr10_+_61685241 | 0.03 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr9_+_2202511 | 0.03 |
ENSRNOT00000017556
|
Satb1
|
SATB homeobox 1 |
| chr5_+_148066062 | 0.03 |
ENSRNOT00000084118
|
AABR07050006.1
|
|
| chr1_+_61374379 | 0.02 |
ENSRNOT00000015343
|
Zfp53
|
zinc finger protein 53 |
| chr8_+_39878955 | 0.02 |
ENSRNOT00000060300
|
LOC100911068
|
roundabout homolog 4-like |
| chr4_+_152449270 | 0.02 |
ENSRNOT00000076733
|
Rad52
|
RAD52 homolog, DNA repair protein |
| chr10_+_88356615 | 0.02 |
ENSRNOT00000022687
|
Klhl10
|
kelch-like family member 10 |
| chr1_-_213973163 | 0.02 |
ENSRNOT00000024867
|
LOC100911402
|
cell cycle exit and neuronal differentiation protein 1-like |
| chrX_+_68774699 | 0.02 |
ENSRNOT00000081662
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr9_-_93404883 | 0.02 |
ENSRNOT00000025024
|
Nmur1
|
neuromedin U receptor 1 |
| chr5_+_172825072 | 0.02 |
ENSRNOT00000068525
|
Cfap74
|
cilia and flagella associated protein 74 |
| chr2_-_210282352 | 0.02 |
ENSRNOT00000075653
|
Slc6a17
|
solute carrier family 6 member 17 |
| chr18_-_63185510 | 0.02 |
ENSRNOT00000024632
|
Afg3l2
|
AFG3 like matrix AAA peptidase subunit 2 |
| chr4_-_56438465 | 0.02 |
ENSRNOT00000064007
|
Rbm28
|
RNA binding motif protein 28 |
| chr1_-_126211439 | 0.02 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr7_+_53630621 | 0.02 |
ENSRNOT00000067011
ENSRNOT00000080598 |
Csrp2
|
cysteine and glycine-rich protein 2 |
| chr6_-_107678156 | 0.02 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr18_-_1946840 | 0.02 |
ENSRNOT00000041878
|
Abhd3
|
abhydrolase domain containing 3 |
| chr4_+_7260575 | 0.02 |
ENSRNOT00000064893
|
Fastk
|
Fas-activated serine/threonine kinase |
| chr20_-_14545772 | 0.02 |
ENSRNOT00000001766
|
Bcr
|
BCR, RhoGEF and GTPase activating protein |
| chr8_+_82380757 | 0.02 |
ENSRNOT00000013512
|
Leo1
|
LEO1 homolog, Paf1/RNA polymerase II complex component |
| chr20_-_5618254 | 0.02 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr7_+_118685181 | 0.02 |
ENSRNOT00000068221
|
Apol3
|
apolipoprotein L, 3 |
| chr10_-_15155412 | 0.02 |
ENSRNOT00000026536
|
Haghl
|
hydroxyacylglutathione hydrolase-like |
| chr8_+_36125999 | 0.02 |
ENSRNOT00000013036
ENSRNOT00000088046 |
Kirrel3
|
kin of IRRE like 3 (Drosophila) |
| chr17_+_45247776 | 0.02 |
ENSRNOT00000081063
ENSRNOT00000080655 |
Zkscan3
|
zinc finger with KRAB and SCAN domains 3 |
| chr8_-_47404010 | 0.02 |
ENSRNOT00000038647
|
Tmem136
|
transmembrane protein 136 |
| chr8_+_127171537 | 0.02 |
ENSRNOT00000078625
ENSRNOT00000051290 ENSRNOT00000087753 |
Golga4
|
golgin A4 |
| chr11_+_61531571 | 0.02 |
ENSRNOT00000093467
ENSRNOT00000002727 |
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr2_+_208420206 | 0.02 |
ENSRNOT00000090500
ENSRNOT00000076065 ENSRNOT00000073644 ENSRNOT00000086279 |
LOC100911453
Wdr77
|
methylosome protein 50-like WD repeat domain 77 |
| chr10_+_81693770 | 0.02 |
ENSRNOT00000003777
ENSRNOT00000085681 |
Spag9
|
sperm associated antigen 9 |
| chr3_-_2727616 | 0.01 |
ENSRNOT00000061904
|
C8g
|
complement C8 gamma chain |
| chr3_+_47453821 | 0.01 |
ENSRNOT00000081682
|
Tank
|
TRAF family member-associated NFKB activator |
| chr4_-_56438142 | 0.01 |
ENSRNOT00000077079
|
Rbm28
|
RNA binding motif protein 28 |
| chr18_-_31430973 | 0.01 |
ENSRNOT00000026065
|
Pcdh12
|
protocadherin 12 |
| chr2_-_238803024 | 0.01 |
ENSRNOT00000046775
|
Tet2
|
tet methylcytosine dioxygenase 2 |
| chr4_-_130659697 | 0.01 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr6_+_2216623 | 0.01 |
ENSRNOT00000008045
|
Rmdn2
|
regulator of microtubule dynamics 2 |
| chr16_+_1979191 | 0.01 |
ENSRNOT00000014382
|
Ppif
|
peptidylprolyl isomerase F |
| chr1_+_80954858 | 0.01 |
ENSRNOT00000030440
|
Zfp112
|
zinc finger protein 112 |
| chr10_+_14248399 | 0.01 |
ENSRNOT00000077689
|
Spsb3
|
splA/ryanodine receptor domain and SOCS box containing 3 |
| chr13_+_100817359 | 0.01 |
ENSRNOT00000004330
|
Tp53bp2
|
tumor protein p53 binding protein, 2 |
| chrX_-_40086870 | 0.01 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr4_-_77706994 | 0.01 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr13_-_110678389 | 0.01 |
ENSRNOT00000082906
|
AABR07022168.1
|
|
| chr3_+_125320680 | 0.01 |
ENSRNOT00000028891
|
LOC681292
|
hypothetical protein LOC681292 |
| chr8_+_48094673 | 0.01 |
ENSRNOT00000008614
|
Nectin1
|
nectin cell adhesion molecule 1 |
| chr18_+_28653592 | 0.01 |
ENSRNOT00000085120
ENSRNOT00000045795 |
Cxxc5
|
CXXC finger protein 5 |
| chr14_+_81362618 | 0.01 |
ENSRNOT00000017386
|
Mfsd10
|
major facilitator superfamily domain containing 10 |
| chr20_+_27352276 | 0.01 |
ENSRNOT00000076534
|
Rn50_20_0292.1
|
|
| chr5_+_25168295 | 0.01 |
ENSRNOT00000020245
|
Fsbp
|
fibrinogen silencer binding protein |
| chr3_+_154905141 | 0.01 |
ENSRNOT00000088748
ENSRNOT00000020556 |
Ralgapb
|
Ral GTPase activating protein non-catalytic beta subunit |
| chrX_+_71342775 | 0.01 |
ENSRNOT00000004888
|
Itgb1bp2
|
integrin subunit beta 1 binding protein 2 |
| chr16_+_3293599 | 0.01 |
ENSRNOT00000081999
ENSRNOT00000047118 ENSRNOT00000020427 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr1_-_134870255 | 0.01 |
ENSRNOT00000055829
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr5_-_129113132 | 0.01 |
ENSRNOT00000080048
|
Rnf11l1
|
ring finger protein 11-like 1 |
| chr9_-_14550625 | 0.01 |
ENSRNOT00000078227
|
Oard1
|
O-acyl-ADP-ribose deacylase 1 |
| chr17_+_6840463 | 0.01 |
ENSRNOT00000061233
|
Ubqln1
|
ubiquilin 1 |
| chr18_-_3555711 | 0.01 |
ENSRNOT00000029274
|
Tmem241
|
transmembrane protein 241 |
| chr7_-_117288018 | 0.01 |
ENSRNOT00000091285
|
Plec
|
plectin |
| chr19_-_10653800 | 0.01 |
ENSRNOT00000022128
|
Cx3cl1
|
C-X3-C motif chemokine ligand 1 |
| chrX_-_78911601 | 0.01 |
ENSRNOT00000003188
|
RGD1566265
|
similar to RIKEN cDNA 2610002M06 |
| chr15_-_37030240 | 0.01 |
ENSRNOT00000028219
ENSRNOT00000088037 |
Pspc1
|
paraspeckle component 1 |
| chr20_-_4817146 | 0.01 |
ENSRNOT00000080174
|
Ddx39b
|
DExD-box helicase 39B |
| chr8_-_116965396 | 0.01 |
ENSRNOT00000042528
|
Bsn
|
bassoon (presynaptic cytomatrix protein) |
| chr1_-_170404056 | 0.00 |
ENSRNOT00000024402
|
Apbb1
|
amyloid beta precursor protein binding family B member 1 |
| chr18_-_63488027 | 0.00 |
ENSRNOT00000023763
|
Ptpn2
|
protein tyrosine phosphatase, non-receptor type 2 |
| chr8_+_71167305 | 0.00 |
ENSRNOT00000021337
|
Rbpms2
|
RNA binding protein with multiple splicing 2 |
| chr2_+_252305874 | 0.00 |
ENSRNOT00000020972
|
Ctbs
|
chitobiase |
| chr13_+_34610684 | 0.00 |
ENSRNOT00000093019
ENSRNOT00000003280 |
Tfcp2l1
|
transcription factor CP2-like 1 |
| chr3_+_113257688 | 0.00 |
ENSRNOT00000019320
|
Map1a
|
microtubule-associated protein 1A |
| chr19_+_9895121 | 0.00 |
ENSRNOT00000033953
|
Prss54
|
protease, serine, 54 |
| chr17_-_15094805 | 0.00 |
ENSRNOT00000044073
|
Fbxw17
|
F-box and WD-40 domain protein 17 |
| chr15_-_4057104 | 0.00 |
ENSRNOT00000084350
ENSRNOT00000012270 |
Sec24c
|
SEC24 homolog C, COPII coat complex component |
| chr9_-_99659425 | 0.00 |
ENSRNOT00000051686
|
Olr1343
|
olfactory receptor 1343 |
| chr4_+_157836912 | 0.00 |
ENSRNOT00000067271
|
Scnn1a
|
sodium channel epithelial 1 alpha subunit |
| chr7_-_136853154 | 0.00 |
ENSRNOT00000087376
|
Nell2
|
neural EGFL like 2 |
| chr1_+_219329574 | 0.00 |
ENSRNOT00000071109
|
Cabp2
|
calcium binding protein 2 |
| chr7_+_143810892 | 0.00 |
ENSRNOT00000016240
|
Znf740
|
zinc finger protein 740 |
| chr12_-_46718355 | 0.00 |
ENSRNOT00000030031
|
Rab35
|
RAB35, member RAS oncogene family |
| chr2_-_111793326 | 0.00 |
ENSRNOT00000092660
|
Nlgn1
|
neuroligin 1 |
| chr5_+_74766636 | 0.00 |
ENSRNOT00000030913
|
Rn50_5_0791.1
|
|
| chr16_-_69242028 | 0.00 |
ENSRNOT00000019002
|
Zfp703
|
zinc finger protein 703 |
| chr15_-_3544685 | 0.00 |
ENSRNOT00000015179
ENSRNOT00000085126 |
Vcl
|
vinculin |
| chr2_+_230186198 | 0.00 |
ENSRNOT00000013049
ENSRNOT00000092954 |
Casp6
|
caspase 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rora
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0032915 | positive regulation of transforming growth factor beta2 production(GO:0032915) |
| 0.0 | 0.1 | GO:0072268 | nephrogenic mesenchyme development(GO:0072076) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0060454 | positive regulation of gastric acid secretion(GO:0060454) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.0 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.0 | 0.0 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.0 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.0 | 0.1 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.0 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.0 | GO:0000172 | ribonuclease MRP complex(GO:0000172) |
| 0.0 | 0.2 | GO:0032591 | dendritic spine membrane(GO:0032591) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.1 | GO:0008349 | MAP kinase kinase kinase kinase activity(GO:0008349) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0000171 | ribonuclease MRP activity(GO:0000171) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |