Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
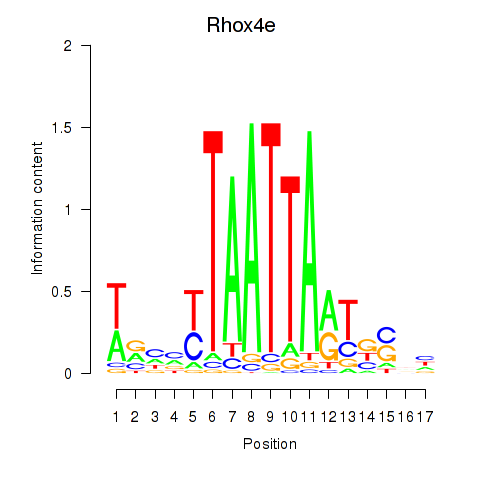
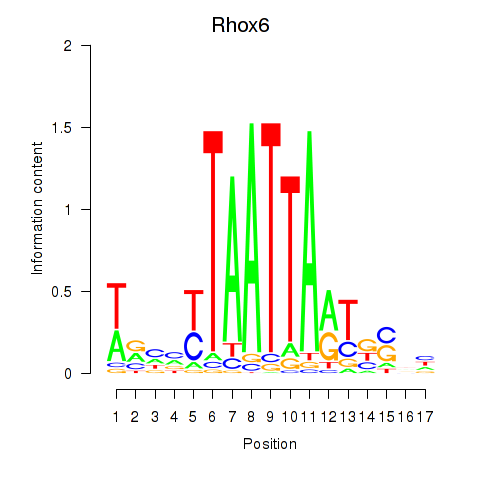
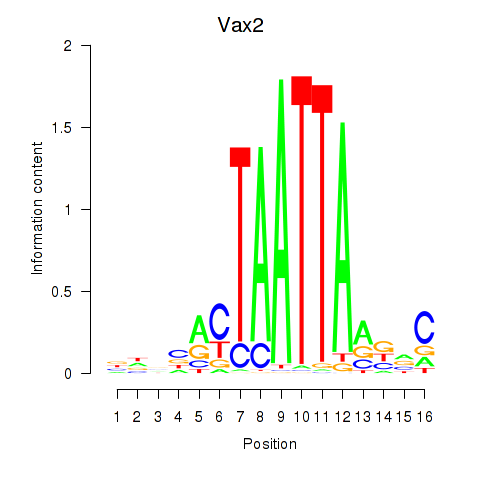
Results for Rhox4e_Rhox6_Vax2
Z-value: 0.29
Transcription factors associated with Rhox4e_Rhox6_Vax2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Vax2
|
ENSRNOG00000013261 | ventral anterior homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Vax2 | rn6_v1_chr4_+_115388839_115388839 | 0.57 | 3.1e-01 | Click! |
Activity profile of Rhox4e_Rhox6_Vax2 motif
Sorted Z-values of Rhox4e_Rhox6_Vax2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_88670430 | 0.45 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr2_+_127525285 | 0.21 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr10_+_61685645 | 0.17 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr1_-_190370499 | 0.17 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chrX_+_15113878 | 0.12 |
ENSRNOT00000007464
|
Wdr13
|
WD repeat domain 13 |
| chr2_+_122877286 | 0.10 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr9_+_71915421 | 0.10 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr1_-_215284548 | 0.09 |
ENSRNOT00000090375
|
Cttn
|
cortactin |
| chr2_-_33025271 | 0.09 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr12_-_35979193 | 0.09 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr2_+_158097843 | 0.08 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr16_+_7035241 | 0.08 |
ENSRNOT00000084977
|
Nek4
|
NIMA-related kinase 4 |
| chr3_-_120076788 | 0.08 |
ENSRNOT00000047158
|
Kcnip3
|
potassium voltage-gated channel interacting protein 3 |
| chr12_+_49665282 | 0.08 |
ENSRNOT00000086397
|
Grk3
|
G protein-coupled receptor kinase 3 |
| chr14_+_34446616 | 0.08 |
ENSRNOT00000002976
|
Clock
|
clock circadian regulator |
| chr14_+_6221085 | 0.07 |
ENSRNOT00000068215
|
AABR07014259.1
|
|
| chr6_+_104291071 | 0.07 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr10_-_10881844 | 0.07 |
ENSRNOT00000087118
ENSRNOT00000004416 |
Mgrn1
|
mahogunin ring finger 1 |
| chr19_+_25043680 | 0.07 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr4_+_158088505 | 0.06 |
ENSRNOT00000026643
|
Vwf
|
von Willebrand factor |
| chr4_+_148782479 | 0.06 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr10_-_106883423 | 0.06 |
ENSRNOT00000071406
|
LOC688286
|
similar to threonine aldolase 1 |
| chr1_+_80279706 | 0.06 |
ENSRNOT00000047105
|
Ppp1r13l
|
protein phosphatase 1, regulatory subunit 13 like |
| chr1_+_87224677 | 0.06 |
ENSRNOT00000028070
|
Ppp1r14a
|
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr1_-_217631862 | 0.05 |
ENSRNOT00000078514
ENSRNOT00000079805 |
Cttn
|
cortactin |
| chr2_-_185852759 | 0.05 |
ENSRNOT00000049461
|
Mab21l2
|
mab-21 like 2 |
| chr7_-_120770435 | 0.05 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chrX_+_74200972 | 0.05 |
ENSRNOT00000076956
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr1_+_88113445 | 0.05 |
ENSRNOT00000056955
|
Ggn
|
gametogenetin |
| chr18_-_55891710 | 0.05 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr1_-_215033460 | 0.05 |
ENSRNOT00000044565
|
Dusp8
|
dual specificity phosphatase 8 |
| chr5_+_139790395 | 0.04 |
ENSRNOT00000015033
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr10_-_65502936 | 0.04 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chrX_+_144994139 | 0.04 |
ENSRNOT00000071783
|
LOC102546596
|
pre-mRNA-splicing factor CWC22 homolog |
| chr9_+_73378057 | 0.04 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr17_-_87826421 | 0.04 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr16_+_7035068 | 0.04 |
ENSRNOT00000024296
|
Nek4
|
NIMA-related kinase 4 |
| chr20_-_13994794 | 0.04 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr1_+_141767940 | 0.03 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr10_-_91302155 | 0.03 |
ENSRNOT00000004371
|
Spata32
|
spermatogenesis associated 32 |
| chr8_+_16287804 | 0.03 |
ENSRNOT00000024691
|
Vkorc1l1
|
vitamin K epoxide reductase complex, subunit 1-like 1 |
| chr4_+_88694583 | 0.03 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr4_-_18396035 | 0.03 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr10_+_43601689 | 0.03 |
ENSRNOT00000029238
|
Mrpl22
|
mitochondrial ribosomal protein L22 |
| chr10_-_67401836 | 0.03 |
ENSRNOT00000073071
|
Crlf3
|
cytokine receptor-like factor 3 |
| chr8_+_40009691 | 0.03 |
ENSRNOT00000042679
|
Vsig2
|
V-set and immunoglobulin domain containing 2 |
| chr4_+_155321553 | 0.03 |
ENSRNOT00000089614
|
Mfap5
|
microfibrillar associated protein 5 |
| chr10_+_106264434 | 0.03 |
ENSRNOT00000003891
|
Sept9
|
septin 9 |
| chr4_+_180062799 | 0.03 |
ENSRNOT00000021623
|
Rassf8
|
Ras association domain family member 8 |
| chr3_-_141411170 | 0.03 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr1_+_8310577 | 0.03 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr17_-_14627937 | 0.03 |
ENSRNOT00000020532
|
NEWGENE_1308171
|
osteoglycin |
| chr20_-_32501722 | 0.03 |
ENSRNOT00000000448
|
Zufsp
|
zinc finger with UFM1-specific peptidase domain |
| chrX_+_6273733 | 0.03 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr4_-_168656673 | 0.03 |
ENSRNOT00000009341
|
Gpr19
|
G protein-coupled receptor 19 |
| chr8_-_83280888 | 0.03 |
ENSRNOT00000052341
|
Gfral
|
GDNF family receptor alpha like |
| chr3_+_56125924 | 0.03 |
ENSRNOT00000011380
|
Ubr3
|
ubiquitin protein ligase E3 component n-recognin 3 |
| chr10_-_74679858 | 0.02 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr12_-_23727535 | 0.02 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr2_+_148765412 | 0.02 |
ENSRNOT00000018248
ENSRNOT00000084662 |
Selenot
|
selenoprotein T |
| chr5_+_187312 | 0.02 |
ENSRNOT00000030857
|
LOC102557117
|
zinc finger protein 120-like |
| chr5_-_147412705 | 0.02 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr6_+_104291340 | 0.02 |
ENSRNOT00000089313
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr17_+_63635086 | 0.02 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr11_-_62067655 | 0.02 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr2_-_35870578 | 0.02 |
ENSRNOT00000066069
|
Rnf180
|
ring finger protein 180 |
| chr10_+_29033989 | 0.02 |
ENSRNOT00000088322
|
Slu7
|
SLU7 homolog, splicing factor |
| chr10_+_103206014 | 0.02 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chr19_+_39229754 | 0.02 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr8_+_71914867 | 0.02 |
ENSRNOT00000023372
|
Dapk2
|
death-associated protein kinase 2 |
| chr6_-_104290579 | 0.02 |
ENSRNOT00000066014
|
Erh
|
enhancer of rudimentary homolog (Drosophila) |
| chr5_-_22765429 | 0.02 |
ENSRNOT00000079432
|
Asph
|
aspartate-beta-hydroxylase |
| chr3_+_148654668 | 0.02 |
ENSRNOT00000081370
|
Tm9sf4
|
transmembrane 9 superfamily member 4 |
| chrX_-_104932508 | 0.02 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chr8_-_7426611 | 0.02 |
ENSRNOT00000031492
|
Arhgap42
|
Rho GTPase activating protein 42 |
| chr2_+_145174876 | 0.02 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr16_+_73584521 | 0.02 |
ENSRNOT00000024301
|
Gins4
|
GINS complex subunit 4 |
| chr18_+_14757679 | 0.02 |
ENSRNOT00000071364
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr7_-_14364178 | 0.02 |
ENSRNOT00000090673
|
Akap8l
|
A-kinase anchoring protein 8 like |
| chr5_+_28485619 | 0.02 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr7_+_12022285 | 0.02 |
ENSRNOT00000024080
|
Rexo1
|
RNA exonuclease 1 homolog |
| chr4_-_157304653 | 0.02 |
ENSRNOT00000051613
|
Lrrc23
|
leucine rich repeat containing 23 |
| chr9_+_20241062 | 0.02 |
ENSRNOT00000071593
|
LOC100911585
|
leucine-rich repeat-containing protein 23-like |
| chr3_+_159368273 | 0.02 |
ENSRNOT00000041688
|
Sgk2
|
serum/glucocorticoid regulated kinase 2 |
| chr17_-_22678253 | 0.02 |
ENSRNOT00000083122
|
LOC100362172
|
LRRGT00112-like |
| chr18_+_87580424 | 0.02 |
ENSRNOT00000070984
|
Tmx3
|
thioredoxin-related transmembrane protein 3 |
| chr1_-_198450047 | 0.02 |
ENSRNOT00000027360
|
Mvp
|
major vault protein |
| chr18_-_43945273 | 0.02 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr7_-_15073052 | 0.02 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr10_-_47546345 | 0.01 |
ENSRNOT00000077461
|
Aldh3a2
|
aldehyde dehydrogenase 3 family, member A2 |
| chr8_-_39460844 | 0.01 |
ENSRNOT00000048875
|
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr1_+_13595295 | 0.01 |
ENSRNOT00000079250
|
Nhsl1
|
NHS-like 1 |
| chr14_-_77810147 | 0.01 |
ENSRNOT00000035427
|
Cytl1
|
cytokine like 1 |
| chr14_-_16903242 | 0.01 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr3_+_8547349 | 0.01 |
ENSRNOT00000079108
ENSRNOT00000091697 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr5_+_122100099 | 0.01 |
ENSRNOT00000007738
|
Pde4b
|
phosphodiesterase 4B |
| chr5_-_16799776 | 0.01 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr1_-_48825364 | 0.01 |
ENSRNOT00000024213
|
Agpat4
|
1-acylglycerol-3-phosphate O-acyltransferase 4 |
| chr8_-_102149912 | 0.01 |
ENSRNOT00000011263
|
RGD1309079
|
similar to Ab2-095 |
| chr3_-_15278645 | 0.01 |
ENSRNOT00000032204
|
Ttll11
|
tubulin tyrosine ligase like11 |
| chr6_-_44363915 | 0.01 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr7_+_2752680 | 0.01 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr5_+_153269526 | 0.01 |
ENSRNOT00000091494
|
Syf2
|
SYF2 pre-mRNA-splicing factor |
| chr1_-_67065797 | 0.01 |
ENSRNOT00000048152
|
Vom1r46
|
vomeronasal 1 receptor 46 |
| chr16_-_83084975 | 0.01 |
ENSRNOT00000017947
|
Arhgef7
|
Rho guanine nucleotide exchange factor 7 |
| chr11_-_86303453 | 0.01 |
ENSRNOT00000071453
|
LOC498122
|
similar to CG15908-PA |
| chr19_-_11263980 | 0.01 |
ENSRNOT00000025086
|
Nup93
|
nucleoporin 93 |
| chr6_-_26241337 | 0.01 |
ENSRNOT00000006525
|
Slc4a1ap
|
solute carrier family 4 member 1 adaptor protein |
| chr2_+_240642847 | 0.01 |
ENSRNOT00000079694
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chr2_+_188583664 | 0.01 |
ENSRNOT00000045477
|
Dpm3
|
dolichyl-phosphate mannosyltransferase subunit 3 |
| chrX_+_74205842 | 0.01 |
ENSRNOT00000077003
|
Chic1
|
cysteine-rich hydrophobic domain 1 |
| chr18_-_16543992 | 0.01 |
ENSRNOT00000036306
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr10_-_94576512 | 0.01 |
ENSRNOT00000035474
|
Icam2
|
intercellular adhesion molecule 2 |
| chr12_+_24761210 | 0.01 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr18_+_4084228 | 0.01 |
ENSRNOT00000071392
|
Cabyr
|
calcium binding tyrosine phosphorylation regulated |
| chr2_+_86891092 | 0.01 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr16_+_39144972 | 0.01 |
ENSRNOT00000086728
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr7_-_28711761 | 0.01 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr5_-_127878792 | 0.01 |
ENSRNOT00000014436
|
Zyg11b
|
zyg-11 family member B, cell cycle regulator |
| chr5_+_154294806 | 0.01 |
ENSRNOT00000012853
|
Hmgcl
|
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr3_+_2591331 | 0.01 |
ENSRNOT00000017466
|
Sapcd2
|
suppressor APC domain containing 2 |
| chr2_+_80269661 | 0.01 |
ENSRNOT00000015975
|
AABR07008940.1
|
|
| chr10_-_97754445 | 0.01 |
ENSRNOT00000088215
|
Slc16a6
|
solute carrier family 16, member 6 |
| chr6_+_26241672 | 0.01 |
ENSRNOT00000006543
|
Supt7l
|
SPT7-like STAGA complex gamma subunit |
| chr11_+_46737892 | 0.01 |
ENSRNOT00000052182
|
AABR07033979.1
|
|
| chr2_-_178521038 | 0.01 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr9_+_88493593 | 0.01 |
ENSRNOT00000020712
|
LOC102556337
|
mitochondrial fission factor-like |
| chr10_-_87286387 | 0.01 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr16_+_71889235 | 0.01 |
ENSRNOT00000038266
|
Adam32
|
ADAM metallopeptidase domain 32 |
| chr1_-_15374850 | 0.01 |
ENSRNOT00000016728
|
Pex7
|
peroxisomal biogenesis factor 7 |
| chr1_-_252461461 | 0.01 |
ENSRNOT00000026093
|
Ankrd22
|
ankyrin repeat domain 22 |
| chr13_-_50514151 | 0.01 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr5_+_155934490 | 0.01 |
ENSRNOT00000077933
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr9_+_79630604 | 0.01 |
ENSRNOT00000021539
ENSRNOT00000083979 |
Tmem169
|
transmembrane protein 169 |
| chr1_-_241046249 | 0.01 |
ENSRNOT00000067796
ENSRNOT00000081866 |
Smc5
|
structural maintenance of chromosomes 5 |
| chr6_-_114476723 | 0.01 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr1_-_134871167 | 0.01 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr19_+_24329023 | 0.01 |
ENSRNOT00000065558
|
Tbc1d9
|
TBC1 domain family member 9 |
| chr4_-_100252755 | 0.01 |
ENSRNOT00000017301
|
Vamp8
|
vesicle-associated membrane protein 8 |
| chr2_+_51672722 | 0.01 |
ENSRNOT00000016485
|
Fgf10
|
fibroblast growth factor 10 |
| chr19_-_50220455 | 0.01 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr6_+_102215603 | 0.01 |
ENSRNOT00000014257
|
Plekhh1
|
pleckstrin homology, MyTH4 and FERM domain containing H1 |
| chr13_+_84474319 | 0.01 |
ENSRNOT00000031367
ENSRNOT00000072244 ENSRNOT00000072897 ENSRNOT00000064168 ENSRNOT00000074954 ENSRNOT00000073696 |
Ildr2
|
immunoglobulin-like domain containing receptor 2 |
| chr17_+_9736786 | 0.01 |
ENSRNOT00000081920
|
F12
|
coagulation factor XII |
| chr20_-_11721838 | 0.01 |
ENSRNOT00000001636
|
Ube2g2
|
ubiquitin-conjugating enzyme E2G 2 |
| chr20_-_9855443 | 0.01 |
ENSRNOT00000090275
ENSRNOT00000066266 |
Tff3
|
trefoil factor 3 |
| chr5_-_166695134 | 0.01 |
ENSRNOT00000047763
|
Tmem201
|
transmembrane protein 201 |
| chr8_+_33239139 | 0.01 |
ENSRNOT00000011589
|
Arhgap32
|
Rho GTPase activating protein 32 |
| chr7_+_26256459 | 0.01 |
ENSRNOT00000010986
|
Appl2
|
adaptor protein, phosphotyrosine interacting with PH domain and leucine zipper 2 |
| chr7_+_44009069 | 0.01 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chr1_+_101554642 | 0.01 |
ENSRNOT00000028474
|
Bcat2
|
branched chain amino acid transaminase 2 |
| chr1_+_229416489 | 0.01 |
ENSRNOT00000028617
|
Olr336
|
olfactory receptor 336 |
| chr10_-_56409017 | 0.01 |
ENSRNOT00000020152
|
Fgf11
|
fibroblast growth factor 11 |
| chr8_-_78096302 | 0.01 |
ENSRNOT00000086185
ENSRNOT00000077999 |
Myzap
|
myocardial zonula adherens protein |
| chr15_-_55209342 | 0.01 |
ENSRNOT00000021752
|
Rb1
|
RB transcriptional corepressor 1 |
| chr11_-_14304603 | 0.01 |
ENSRNOT00000040202
ENSRNOT00000082143 |
Samsn1
|
SAM domain, SH3 domain and nuclear localization signals, 1 |
| chr19_-_29968424 | 0.01 |
ENSRNOT00000024981
|
Inpp4b
|
inositol polyphosphate-4-phosphatase type II B |
| chr5_-_154222702 | 0.01 |
ENSRNOT00000080069
|
Pnrc2
|
proline-rich nuclear receptor coactivator 2 |
| chr1_+_188420461 | 0.01 |
ENSRNOT00000037870
|
Ccp110
|
centriolar coiled-coil protein 110 |
| chr14_+_39964588 | 0.00 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr4_+_180291389 | 0.00 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr1_-_88657845 | 0.00 |
ENSRNOT00000074211
|
LOC100912380
|
calpain small subunit 1-like |
| chr19_+_45938915 | 0.00 |
ENSRNOT00000065508
|
Mon1b
|
MON1 homolog B, secretory trafficking associated |
| chr16_-_48692476 | 0.00 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr3_-_91370167 | 0.00 |
ENSRNOT00000006291
|
Prr5l
|
proline rich 5 like |
| chr8_+_117062884 | 0.00 |
ENSRNOT00000082452
ENSRNOT00000071540 |
Nicn1
|
nicolin 1 |
| chr18_-_26656879 | 0.00 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr12_-_37538403 | 0.00 |
ENSRNOT00000001402
|
Snrnp35
|
small nuclear ribonucleoprotein U11/U12 subunit 35 |
| chr1_-_214067657 | 0.00 |
ENSRNOT00000044390
|
LOC100911881
|
chitinase domain-containing protein 1-like |
| chr18_+_79773608 | 0.00 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr2_-_60657712 | 0.00 |
ENSRNOT00000040348
|
Rai14
|
retinoic acid induced 14 |
| chr15_-_95514259 | 0.00 |
ENSRNOT00000038433
|
Slitrk6
|
SLIT and NTRK-like family, member 6 |
| chr7_-_91538673 | 0.00 |
ENSRNOT00000006209
|
Rad21
|
RAD21 cohesin complex component |
| chr11_-_87403367 | 0.00 |
ENSRNOT00000057751
|
Thap7
|
THAP domain containing 7 |
| chr8_-_84506328 | 0.00 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr7_-_29171783 | 0.00 |
ENSRNOT00000079235
|
Mybpc1
|
myosin binding protein C, slow type |
| chr12_-_5685448 | 0.00 |
ENSRNOT00000077167
|
Fry
|
FRY microtubule binding protein |
| chrX_-_32095867 | 0.00 |
ENSRNOT00000049947
ENSRNOT00000080730 |
Ace2
|
angiotensin I converting enzyme 2 |
| chr9_-_11027506 | 0.00 |
ENSRNOT00000071107
|
Chaf1a
|
chromatin assembly factor 1 subunit A |
| chr4_+_162937774 | 0.00 |
ENSRNOT00000083188
|
Clec2g
|
C-type lectin domain family 2, member G |
| chr13_+_31081804 | 0.00 |
ENSRNOT00000041413
|
Cdh7
|
cadherin 7 |
| chr8_-_33121002 | 0.00 |
ENSRNOT00000047211
|
LOC100362981
|
LRRGT00010-like |
| chr1_+_80141630 | 0.00 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr8_+_117297670 | 0.00 |
ENSRNOT00000082628
|
Qars
|
glutaminyl-tRNA synthetase |
| chr9_+_60883981 | 0.00 |
ENSRNOT00000081002
|
Ccdc150
|
coiled-coil domain containing 150 |
| chr4_+_99823252 | 0.00 |
ENSRNOT00000013587
|
Polr1a
|
RNA polymerase I subunit A |
| chr13_+_67611708 | 0.00 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr2_-_58534211 | 0.00 |
ENSRNOT00000089178
|
Skp2
|
S-phase kinase associated protein 2 |
| chr20_+_3155652 | 0.00 |
ENSRNOT00000042882
|
RT1-S2
|
RT1 class Ib, locus S2 |
| chr14_-_44078897 | 0.00 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr3_+_17889972 | 0.00 |
ENSRNOT00000073021
|
AABR07051611.1
|
|
| chr6_-_23316962 | 0.00 |
ENSRNOT00000065421
|
Clip4
|
CAP-GLY domain containing linker protein family, member 4 |
| chr3_+_3389612 | 0.00 |
ENSRNOT00000041984
|
Rpl8
|
ribosomal protein L8 |
| chr8_+_85059051 | 0.00 |
ENSRNOT00000033196
|
Gclc
|
glutamate-cysteine ligase, catalytic subunit |
| chr3_+_173799833 | 0.00 |
ENSRNOT00000081235
|
Phactr3
|
phosphatase and actin regulator 3 |
| chr5_-_119564846 | 0.00 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chr16_+_39145230 | 0.00 |
ENSRNOT00000092942
|
Adam21
|
ADAM metallopeptidase domain 21 |
| chr14_+_44889287 | 0.00 |
ENSRNOT00000091312
ENSRNOT00000032273 |
Tmem156
|
transmembrane protein 156 |
| chr17_+_2690062 | 0.00 |
ENSRNOT00000024937
|
Olr1653
|
olfactory receptor 1653 |
| chr16_+_23447366 | 0.00 |
ENSRNOT00000068629
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rhox4e_Rhox6_Vax2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.1 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.1 | GO:0043324 | eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) rhodopsin metabolic process(GO:0046154) |
| 0.0 | 0.1 | GO:0006567 | threonine catabolic process(GO:0006567) |
| 0.0 | 0.1 | GO:0098885 | modification of postsynaptic actin cytoskeleton(GO:0098885) |
| 0.0 | 0.2 | GO:0010839 | negative regulation of keratinocyte proliferation(GO:0010839) |
| 0.0 | 0.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.0 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.0 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0098871 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.0 | 0.0 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008732 | threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.0 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0016308 | 1-phosphatidylinositol-4-phosphate 5-kinase activity(GO:0016308) |
| 0.0 | 0.0 | GO:0016900 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |