Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
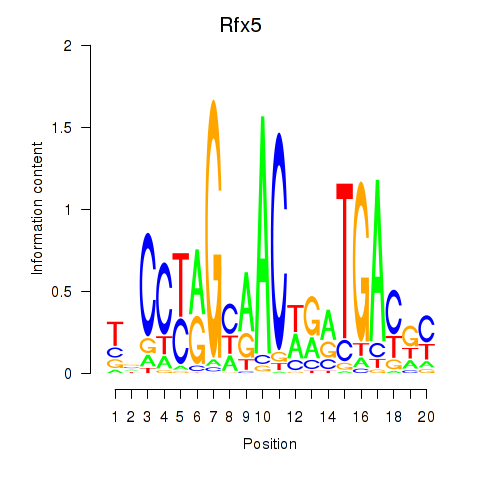
Results for Rfx5
Z-value: 0.58
Transcription factors associated with Rfx5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx5
|
ENSRNOG00000021012 | regulatory factor X5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx5 | rn6_v1_chr2_+_196120580_196120580 | -0.86 | 6.3e-02 | Click! |
Activity profile of Rfx5 motif
Sorted Z-values of Rfx5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_55491152 | 0.24 |
ENSRNOT00000014965
|
LOC100359479
|
rCG58364-like |
| chr20_-_3978845 | 0.24 |
ENSRNOT00000000532
|
Psmb9
|
proteasome subunit beta 9 |
| chr1_-_226526959 | 0.22 |
ENSRNOT00000028003
|
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chr9_-_15274917 | 0.22 |
ENSRNOT00000019650
|
Pgc
|
progastricsin |
| chr20_+_3979035 | 0.20 |
ENSRNOT00000000529
|
Tap1
|
transporter 1, ATP binding cassette subfamily B member |
| chr20_-_5244386 | 0.20 |
ENSRNOT00000070886
|
RT1-DMa
|
RT1 class II, locus DMa |
| chr20_-_5227620 | 0.19 |
ENSRNOT00000086240
|
RT1-DMb
|
RT1 class II, locus DMb |
| chr20_+_3935529 | 0.19 |
ENSRNOT00000072458
|
LOC108348072
|
class II histocompatibility antigen, M alpha chain |
| chr20_-_5234290 | 0.19 |
ENSRNOT00000073474
|
RT1-DMb
|
RT1 class II, locus DMb |
| chr20_+_3945601 | 0.18 |
ENSRNOT00000075342
|
RT1-DMb
|
RT1 class II, locus DMb |
| chr20_+_4178686 | 0.17 |
ENSRNOT00000080641
|
Tesb
|
testis specific basic protein |
| chr10_+_71217966 | 0.15 |
ENSRNOT00000076192
|
Hnf1b
|
HNF1 homeobox B |
| chr20_+_4087618 | 0.15 |
ENSRNOT00000000522
ENSRNOT00000060327 ENSRNOT00000080590 |
RT1-Db1
|
RT1 class II, locus Db1 |
| chr20_+_5414448 | 0.14 |
ENSRNOT00000078972
ENSRNOT00000080900 |
RT1-A1
|
RT1 class Ia, locus A1 |
| chr1_+_142951094 | 0.14 |
ENSRNOT00000077441
|
Slc28a1
|
solute carrier family 28 member 1 |
| chrX_+_131617798 | 0.14 |
ENSRNOT00000074384
|
Prr32
|
proline rich 32 |
| chr18_+_56071478 | 0.14 |
ENSRNOT00000025344
ENSRNOT00000025354 |
Cd74
|
CD74 molecule |
| chr1_+_100473643 | 0.13 |
ENSRNOT00000026379
|
Josd2
|
Josephin domain containing 2 |
| chr20_+_4043741 | 0.13 |
ENSRNOT00000000525
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr19_-_26181449 | 0.12 |
ENSRNOT00000036953
|
LOC685513
|
similar to Guanine nucleotide-binding protein G(I)/G(S)/G(O) gamma-12 subunit precursor |
| chr9_-_121713091 | 0.11 |
ENSRNOT00000073432
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr20_+_5374985 | 0.11 |
ENSRNOT00000052270
|
RT1-A2
|
RT1 class Ia, locus A2 |
| chr10_+_94466523 | 0.10 |
ENSRNOT00000014867
|
Tcam1
|
testicular cell adhesion molecule 1 |
| chr20_-_1818800 | 0.10 |
ENSRNOT00000000990
|
RT1-M3-1
|
RT1 class Ib, locus M3, gene 1 |
| chr19_+_37830917 | 0.09 |
ENSRNOT00000025608
|
Nutf2
|
nuclear transport factor 2 |
| chr3_+_114087287 | 0.09 |
ENSRNOT00000023017
|
B2m
|
beta-2 microglobulin |
| chr2_-_89310946 | 0.08 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr10_+_49020454 | 0.07 |
ENSRNOT00000003402
ENSRNOT00000067379 |
LOC108348055
|
B9 domain-containing protein 1 |
| chr10_+_47785033 | 0.07 |
ENSRNOT00000075073
ENSRNOT00000084944 |
B9d1
|
B9 domain containing 1 |
| chr20_-_4896970 | 0.07 |
ENSRNOT00000061114
ENSRNOT00000060980 |
RT1-CE7
RT1-CE5
|
RT1 class I, locus CE7 RT1 class I, locus CE5 |
| chr1_-_101085884 | 0.06 |
ENSRNOT00000085352
|
Rcn3
|
reticulocalbin 3 |
| chr1_+_256035866 | 0.06 |
ENSRNOT00000079726
|
Kif11
|
kinesin family member 11 |
| chr2_-_256154584 | 0.06 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr20_-_157861 | 0.06 |
ENSRNOT00000084461
|
RT1-CE10
|
RT1 class I, locus CE10 |
| chr20_-_3166569 | 0.06 |
ENSRNOT00000091390
|
AABR07044362.1
|
|
| chr7_+_54859104 | 0.05 |
ENSRNOT00000087341
|
Caps2
|
calcyphosine 2 |
| chr19_-_25955371 | 0.05 |
ENSRNOT00000004042
ENSRNOT00000084123 |
Rad23a
|
RAD23 homolog A, nucleotide excision repair protein |
| chr13_-_36290531 | 0.05 |
ENSRNOT00000071388
|
Steap3
|
STEAP3 metalloreductase |
| chr10_-_14247886 | 0.05 |
ENSRNOT00000020504
|
Nubp2
|
nucleotide binding protein 2 |
| chr20_-_157665 | 0.05 |
ENSRNOT00000048858
ENSRNOT00000079494 |
RT1-CE10
|
RT1 class I, locus CE10 |
| chrX_+_37329779 | 0.05 |
ENSRNOT00000038352
ENSRNOT00000088802 |
Pdha1
|
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr7_+_117420788 | 0.05 |
ENSRNOT00000049198
|
Wdr97
|
WD repeat domain 97 |
| chr8_+_56411911 | 0.05 |
ENSRNOT00000039083
|
LOC100359687
|
mitochondrial ribosomal protein L1-like |
| chr7_+_54859326 | 0.05 |
ENSRNOT00000039141
|
Caps2
|
calcyphosine 2 |
| chr4_-_66380734 | 0.04 |
ENSRNOT00000009194
|
Klrg2
|
killer cell lectin like receptor G2 |
| chr13_-_71282802 | 0.04 |
ENSRNOT00000025544
|
Rgsl2h
|
regulator of G-protein signaling like 2 homolog (mouse) |
| chr19_-_37528011 | 0.04 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr3_-_123997277 | 0.04 |
ENSRNOT00000058238
|
RGD1564425
|
similar to Proteasome subunit beta type 3 (Proteasome theta chain) |
| chr10_-_72196437 | 0.03 |
ENSRNOT00000029769
|
Pigw
|
phosphatidylinositol glycan anchor biosynthesis, class W |
| chr13_-_72789841 | 0.03 |
ENSRNOT00000004694
|
Mr1
|
major histocompatibility complex, class I-related |
| chr20_+_4039413 | 0.03 |
ENSRNOT00000082136
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr11_-_67618704 | 0.03 |
ENSRNOT00000042208
|
Ccdc58
|
coiled-coil domain containing 58 |
| chr12_+_37829579 | 0.03 |
ENSRNOT00000084587
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr14_+_17534412 | 0.03 |
ENSRNOT00000079304
ENSRNOT00000046771 |
Cdkl2
|
cyclin dependent kinase like 2 |
| chr5_-_135472116 | 0.03 |
ENSRNOT00000022170
|
Nasp
|
nuclear autoantigenic sperm protein |
| chr1_+_189550354 | 0.02 |
ENSRNOT00000083153
|
Exnef
|
exonuclease NEF-sp |
| chr11_+_47243342 | 0.02 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr10_-_40201992 | 0.02 |
ENSRNOT00000075311
|
Lyrm7
|
LYR motif containing 7 |
| chr1_-_170594168 | 0.01 |
ENSRNOT00000026280
|
Tpp1
|
tripeptidyl peptidase 1 |
| chr2_-_149333597 | 0.01 |
ENSRNOT00000066948
|
P2ry14
|
purinergic receptor P2Y14 |
| chr4_+_181428664 | 0.01 |
ENSRNOT00000002518
|
Rep15
|
RAB15 effector protein |
| chr11_+_67618625 | 0.01 |
ENSRNOT00000087054
ENSRNOT00000043230 ENSRNOT00000041844 |
Fam162a
|
family with sequence similarity 162, member A |
| chr3_+_159823878 | 0.01 |
ENSRNOT00000011748
|
Gdap1l1
|
ganglioside-induced differentiation-associated protein 1-like 1 |
| chr15_-_20032191 | 0.00 |
ENSRNOT00000032704
|
Ddhd1
|
DDHD domain containing 1 |
| chr20_-_3401273 | 0.00 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
| chr3_-_72289310 | 0.00 |
ENSRNOT00000038250
|
Rtn4rl2
|
reticulon 4 receptor-like 2 |
| chr7_-_29701586 | 0.00 |
ENSRNOT00000009084
ENSRNOT00000089269 |
Ano4
|
anoctamin 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0002503 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.1 | 0.2 | GO:0002481 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.1 | 0.2 | GO:0046967 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 0.1 | 0.2 | GO:1901423 | response to benzene(GO:1901423) |
| 0.1 | 0.2 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.2 | GO:0035565 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.1 | GO:0002462 | tolerance induction to nonself antigen(GO:0002462) |
| 0.0 | 0.2 | GO:0002343 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.1 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:1904823 | purine nucleobase transmembrane transport(GO:1904823) |
| 0.0 | 0.1 | GO:0002906 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.1 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0019885 | antigen processing and presentation of endogenous peptide antigen via MHC class I(GO:0019885) |
| 0.0 | 0.1 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.0 | 0.0 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0046602 | regulation of mitotic centrosome separation(GO:0046602) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.6 | GO:0042611 | MHC protein complex(GO:0042611) |
| 0.0 | 0.2 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0044613 | nuclear pore central transport channel(GO:0044613) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0046980 | peptide antigen-transporting ATPase activity(GO:0015433) tapasin binding(GO:0046980) |
| 0.0 | 1.0 | GO:0023026 | MHC class II protein complex binding(GO:0023026) |
| 0.0 | 0.1 | GO:0015389 | pyrimidine- and adenine-specific:sodium symporter activity(GO:0015389) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0046979 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0030881 | beta-2-microglobulin binding(GO:0030881) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.0 | 0.2 | GO:1990405 | protein antigen binding(GO:1990405) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.2 | GO:0070628 | proteasome binding(GO:0070628) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |