Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
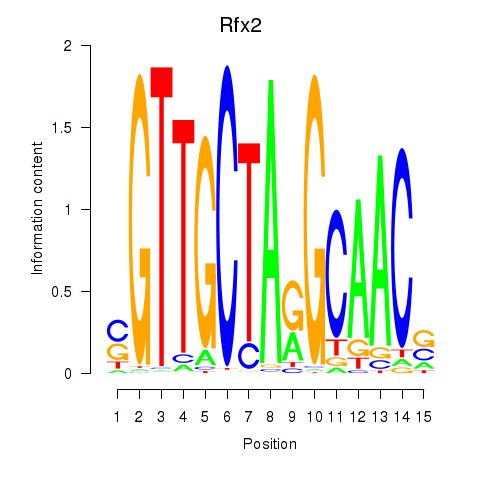
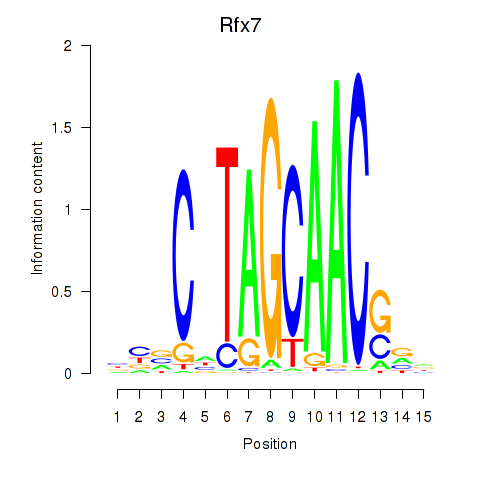
Results for Rfx2_Rfx7
Z-value: 1.59
Transcription factors associated with Rfx2_Rfx7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rfx2
|
ENSRNOG00000045846 | regulatory factor X2 |
|
Rfx7
|
ENSRNOG00000060367 | regulatory factor X, 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rfx2 | rn6_v1_chr9_+_10216205_10216293 | -0.99 | 9.2e-04 | Click! |
| Rfx7 | rn6_v1_chr8_+_79159370_79159370 | -0.96 | 8.5e-03 | Click! |
Activity profile of Rfx2_Rfx7 motif
Sorted Z-values of Rfx2_Rfx7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_100473643 | 1.08 |
ENSRNOT00000026379
|
Josd2
|
Josephin domain containing 2 |
| chr20_+_14577166 | 0.99 |
ENSRNOT00000085583
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr5_-_147817852 | 0.92 |
ENSRNOT00000071940
|
Dcdc2b
|
doublecortin domain containing 2B |
| chr1_-_226526959 | 0.92 |
ENSRNOT00000028003
|
Ppp1r32
|
protein phosphatase 1, regulatory subunit 32 |
| chrX_+_63520991 | 0.86 |
ENSRNOT00000071590
|
Apoo
|
apolipoprotein O |
| chr10_-_84976170 | 0.84 |
ENSRNOT00000013542
|
Lrrc46
|
leucine rich repeat containing 46 |
| chr6_+_52702544 | 0.78 |
ENSRNOT00000014252
|
Efcab10
|
EF-hand calcium binding domain 10 |
| chr19_-_53754602 | 0.72 |
ENSRNOT00000035651
|
Fam92b
|
family with sequence similarity 92, member B |
| chr1_+_100891866 | 0.71 |
ENSRNOT00000086890
ENSRNOT00000083336 |
Fuz
|
fuzzy planar cell polarity protein |
| chr11_+_84094520 | 0.70 |
ENSRNOT00000046642
|
Rps15al2
|
ribosomal protein S15A-like 2 |
| chr17_-_88095729 | 0.66 |
ENSRNOT00000025140
|
Enkur
|
enkurin, TRPC channel interacting protein |
| chr16_+_56248331 | 0.63 |
ENSRNOT00000085300
|
Tusc3
|
tumor suppressor candidate 3 |
| chr17_+_85914410 | 0.62 |
ENSRNOT00000075188
|
Armc3
|
armadillo repeat containing 3 |
| chr1_-_145931583 | 0.60 |
ENSRNOT00000016433
|
Cfap161
|
cilia and flagella associated protein 161 |
| chr7_+_54213319 | 0.59 |
ENSRNOT00000005286
|
Nap1l1
|
nucleosome assembly protein 1-like 1 |
| chr6_-_27323992 | 0.57 |
ENSRNOT00000013198
|
RGD1559683
|
similar to RIKEN cDNA 1700001C02 |
| chr8_+_79638696 | 0.55 |
ENSRNOT00000085959
|
Dyx1c1
|
dyslexia susceptibility 1 candidate 1 |
| chr10_-_10767389 | 0.54 |
ENSRNOT00000066754
|
Smim22
|
small integral membrane protein 22 |
| chr10_-_44746549 | 0.52 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr5_-_136112344 | 0.51 |
ENSRNOT00000050195
|
RGD1563714
|
RGD1563714 |
| chr6_+_135856218 | 0.51 |
ENSRNOT00000066715
|
RGD1560608
|
similar to novel protein |
| chr8_+_79054237 | 0.50 |
ENSRNOT00000077613
|
Mns1
|
meiosis-specific nuclear structural 1 |
| chr15_-_34352673 | 0.47 |
ENSRNOT00000064916
|
Nedd8
|
neural precursor cell expressed, developmentally down-regulated 8 |
| chr20_-_14573519 | 0.44 |
ENSRNOT00000001772
|
Rab36
|
RAB36, member RAS oncogene family |
| chr6_-_59950586 | 0.43 |
ENSRNOT00000005800
|
Arl4a
|
ADP-ribosylation factor like GTPase 4A |
| chr20_-_12938891 | 0.43 |
ENSRNOT00000017141
|
RGD1564149
|
similar to Protein C21orf58 |
| chr10_+_88459490 | 0.43 |
ENSRNOT00000080231
ENSRNOT00000067559 |
Ttc25
|
tetratricopeptide repeat domain 25 |
| chr2_-_251970768 | 0.42 |
ENSRNOT00000020141
|
Wdr63
|
WD repeat domain 63 |
| chr1_+_72732668 | 0.41 |
ENSRNOT00000024251
|
Hspbp1
|
HSPA (heat shock 70) binding protein, cytoplasmic cochaperone 1 |
| chr10_+_14828597 | 0.41 |
ENSRNOT00000025434
|
Tekt4
|
tektin 4 |
| chr7_-_115981731 | 0.40 |
ENSRNOT00000036549
|
RGD1308195
|
similar to secreted Ly6/uPAR related protein 2 |
| chr19_+_44164935 | 0.38 |
ENSRNOT00000048998
|
Gabarapl2
|
GABA type A receptor associated protein like 2 |
| chr5_+_136112417 | 0.37 |
ENSRNOT00000025990
|
Tmem53
|
transmembrane protein 53 |
| chr16_-_14348046 | 0.37 |
ENSRNOT00000018630
|
Cdhr1
|
cadherin-related family member 1 |
| chr19_+_49016891 | 0.37 |
ENSRNOT00000016713
|
Dynlrb2
|
dynein light chain roadblock-type 2 |
| chr12_+_46042413 | 0.36 |
ENSRNOT00000046882
|
Ccdc60
|
coiled-coil domain containing 60 |
| chr10_+_49472460 | 0.35 |
ENSRNOT00000038276
|
Tekt3
|
tektin 3 |
| chr3_-_7051953 | 0.35 |
ENSRNOT00000013473
|
RGD1306233
|
similar to hypothetical protein MGC29761 |
| chr10_+_103266296 | 0.35 |
ENSRNOT00000038852
|
Dnai2
|
dynein, axonemal, intermediate chain 2 |
| chr1_-_154111725 | 0.35 |
ENSRNOT00000055488
|
Ccdc81
|
coiled-coil domain containing 81 |
| chr16_+_20400577 | 0.34 |
ENSRNOT00000037756
|
Mast3
|
microtubule associated serine/threonine kinase 3 |
| chr14_+_91557601 | 0.34 |
ENSRNOT00000038733
|
RGD1309870
|
hypothetical LOC289778 |
| chr20_+_33945829 | 0.34 |
ENSRNOT00000064063
|
LOC103689994
|
radial spoke head protein 4 homolog A |
| chr20_+_32450733 | 0.34 |
ENSRNOT00000036449
|
Rsph4a
|
radial spoke head 4 homolog A |
| chr17_+_78817529 | 0.33 |
ENSRNOT00000021918
|
Meig1
|
meiosis/spermiogenesis associated 1 |
| chr1_-_199037267 | 0.32 |
ENSRNOT00000078779
|
Ccdc189
|
coiled-coil domain containing 189 |
| chr12_+_19714324 | 0.32 |
ENSRNOT00000072303
|
RGD1559588
|
similar to cell surface receptor FDFACT |
| chr1_-_101803200 | 0.32 |
ENSRNOT00000028588
|
Cyth2
|
cytohesin 2 |
| chr18_-_27374603 | 0.32 |
ENSRNOT00000027244
|
Nme5
|
NME/NM23 family member 5 |
| chr2_-_208633945 | 0.30 |
ENSRNOT00000049155
|
Pifo
|
primary cilia formation |
| chr20_-_10680283 | 0.30 |
ENSRNOT00000001579
|
Sik1
|
salt-inducible kinase 1 |
| chr13_+_83681322 | 0.29 |
ENSRNOT00000004206
|
Mpc2
|
mitochondrial pyruvate carrier 2 |
| chr1_+_84044551 | 0.29 |
ENSRNOT00000028290
|
Coq8b
|
coenzyme Q8B |
| chr8_-_23014499 | 0.29 |
ENSRNOT00000017820
|
Ccdc151
|
coiled-coil domain containing 151 |
| chr6_+_137386422 | 0.29 |
ENSRNOT00000018568
|
RGD1307315
|
LOC362793 |
| chr10_-_57671080 | 0.28 |
ENSRNOT00000082511
|
LOC691995
|
hypothetical protein LOC691995 |
| chr6_+_99817431 | 0.28 |
ENSRNOT00000009621
|
Churc1
|
churchill domain containing 1 |
| chrX_+_76042239 | 0.28 |
ENSRNOT00000003667
|
Pbdc1
|
polysaccharide biosynthesis domain containing 1 |
| chr1_+_137014272 | 0.28 |
ENSRNOT00000014802
|
Akap13
|
A-kinase anchoring protein 13 |
| chr5_+_118574801 | 0.28 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr7_+_121841855 | 0.28 |
ENSRNOT00000024673
|
Grap2
|
GRB2-related adaptor protein 2 |
| chr19_+_10167797 | 0.28 |
ENSRNOT00000085440
|
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr7_-_123156558 | 0.27 |
ENSRNOT00000005943
|
Polr3h
|
RNA polymerase III subunit H |
| chr1_+_214182830 | 0.27 |
ENSRNOT00000022867
|
Lrrc56
|
leucine rich repeat containing 56 |
| chr2_+_252263386 | 0.27 |
ENSRNOT00000092913
ENSRNOT00000084034 ENSRNOT00000041186 ENSRNOT00000092931 |
Ssx2ip
|
SSX family member 2 interacting protein |
| chr7_+_126028350 | 0.26 |
ENSRNOT00000042412
|
Ribc2
|
RIB43A domain with coiled-coils 2 |
| chr6_-_11063073 | 0.26 |
ENSRNOT00000086010
ENSRNOT00000061746 |
LOC681766
|
hypothetical protein LOC681766 |
| chr8_-_112594261 | 0.26 |
ENSRNOT00000015240
|
Uba5
|
ubiquitin-like modifier activating enzyme 5 |
| chr10_+_106065712 | 0.25 |
ENSRNOT00000003690
|
Sec14l1
|
SEC14-like lipid binding 1 |
| chr10_-_95934345 | 0.25 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr4_+_171250818 | 0.25 |
ENSRNOT00000040576
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr10_+_14062331 | 0.24 |
ENSRNOT00000029652
|
Noxo1
|
NADPH oxidase organizer 1 |
| chr4_+_169147243 | 0.24 |
ENSRNOT00000011580
|
Emp1
|
epithelial membrane protein 1 |
| chr4_+_114854458 | 0.24 |
ENSRNOT00000013312
|
LOC500227
|
hypothetical gene supported by BC079424 |
| chr10_+_55169282 | 0.24 |
ENSRNOT00000005423
|
Ccdc42
|
coiled-coil domain containing 42 |
| chr16_-_82439441 | 0.24 |
ENSRNOT00000040315
|
AABR07026539.1
|
|
| chr7_+_11724962 | 0.23 |
ENSRNOT00000026551
|
Lsm7
|
LSM7 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_+_267689328 | 0.23 |
ENSRNOT00000077738
|
Cfap58
|
cilia and flagella associated protein 58 |
| chr7_+_54859104 | 0.23 |
ENSRNOT00000087341
|
Caps2
|
calcyphosine 2 |
| chr2_+_164549455 | 0.23 |
ENSRNOT00000017151
|
Mlf1
|
myeloid leukemia factor 1 |
| chr1_-_72464492 | 0.23 |
ENSRNOT00000068550
|
Nat14
|
N-acetyltransferase 14 |
| chrX_+_156963870 | 0.23 |
ENSRNOT00000077109
|
Pdzd4
|
PDZ domain containing 4 |
| chr5_+_57947716 | 0.23 |
ENSRNOT00000067657
|
Dnai1
|
dynein, axonemal, intermediate chain 1 |
| chr5_+_161889342 | 0.22 |
ENSRNOT00000040481
|
LOC100362684
|
ribosomal protein S20-like |
| chr3_+_171597241 | 0.22 |
ENSRNOT00000029558
|
LOC689618
|
similar to Protein C20orf85 homolog |
| chr8_-_79660439 | 0.22 |
ENSRNOT00000089949
|
Ccpg1os
|
cell cycle progression 1, opposite strand |
| chr1_-_164849749 | 0.22 |
ENSRNOT00000024772
|
Spcs2
|
signal peptidase complex subunit 2 |
| chr6_+_129538982 | 0.22 |
ENSRNOT00000083626
ENSRNOT00000082522 |
Ak7
|
adenylate kinase 7 |
| chr7_-_74901997 | 0.22 |
ENSRNOT00000039378
|
Rgs22
|
regulator of G-protein signaling 22 |
| chr5_-_78324278 | 0.22 |
ENSRNOT00000082642
ENSRNOT00000048904 |
Wdr31
|
WD repeat domain 31 |
| chr10_-_109811323 | 0.22 |
ENSRNOT00000054970
|
Mafg
|
MAF bZIP transcription factor G |
| chr16_-_7082193 | 0.21 |
ENSRNOT00000024447
|
Spcs1
|
signal peptidase complex subunit 1 |
| chr14_+_106393959 | 0.21 |
ENSRNOT00000092168
|
Wdpcp
|
WD repeat containing planar cell polarity effector |
| chr1_+_53220397 | 0.21 |
ENSRNOT00000089989
|
AABR07001573.1
|
|
| chr18_+_72005581 | 0.21 |
ENSRNOT00000072519
|
Zbtb7c
|
zinc finger and BTB domain containing 7C |
| chr9_-_43127887 | 0.21 |
ENSRNOT00000021685
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr4_-_145555748 | 0.21 |
ENSRNOT00000013503
|
Fancd2os
|
FANCD2 opposite strand |
| chr10_+_109107389 | 0.20 |
ENSRNOT00000068437
ENSRNOT00000005687 |
Baiap2
|
BAI1-associated protein 2 |
| chr9_+_27068443 | 0.20 |
ENSRNOT00000017393
|
Efhc1
|
EF-hand domain containing 1 |
| chr7_-_121058029 | 0.20 |
ENSRNOT00000068033
|
Cbx6
|
chromobox 6 |
| chr4_-_159079003 | 0.20 |
ENSRNOT00000026691
|
Kcna5
|
potassium voltage-gated channel subfamily A member 5 |
| chr2_+_174203693 | 0.20 |
ENSRNOT00000073540
|
LOC108350036
|
uncharacterized LOC108350036 |
| chr3_+_164424515 | 0.19 |
ENSRNOT00000083876
|
Cebpb
|
CCAAT/enhancer binding protein beta |
| chr13_-_89745835 | 0.19 |
ENSRNOT00000005352
|
Klhdc9
|
kelch domain containing 9 |
| chr9_+_66335492 | 0.19 |
ENSRNOT00000037555
|
RGD1562029
|
similar to KIAA2012 protein |
| chr18_-_33414236 | 0.19 |
ENSRNOT00000020385
|
Yipf5
|
Yip1 domain family, member 5 |
| chr1_-_37726151 | 0.19 |
ENSRNOT00000071842
|
RGD1308544
|
LOC361192 |
| chr2_-_243475639 | 0.18 |
ENSRNOT00000089222
|
RGD1309170
|
similar to hypothetical protein DKFZp434G072 |
| chr5_-_144345531 | 0.18 |
ENSRNOT00000014721
|
Tekt2
|
tektin 2 |
| chr16_+_18648866 | 0.18 |
ENSRNOT00000014862
|
Dydc1
|
DPY30 domain containing 1 |
| chr4_-_113988246 | 0.18 |
ENSRNOT00000013128
|
LOC103692166
|
WD repeat-containing protein 54 |
| chr1_-_146289465 | 0.18 |
ENSRNOT00000017362
|
Abhd17c
|
abhydrolase domain containing 17C |
| chr1_-_212317244 | 0.18 |
ENSRNOT00000068604
|
LOC103691261
|
sperm flagellar protein 1-like |
| chr20_-_3374344 | 0.17 |
ENSRNOT00000082999
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr2_-_178612470 | 0.17 |
ENSRNOT00000013499
|
Tmem144
|
transmembrane protein 144 |
| chr2_+_181331464 | 0.17 |
ENSRNOT00000017448
|
Map9
|
microtubule-associated protein 9 |
| chr6_-_135112775 | 0.17 |
ENSRNOT00000086310
|
LOC103692716
|
heat shock protein HSP 90-alpha |
| chr13_+_97702097 | 0.17 |
ENSRNOT00000057787
|
LOC103689999
|
saccharopine dehydrogenase-like oxidoreductase |
| chr17_+_44522140 | 0.17 |
ENSRNOT00000080490
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr14_+_86922223 | 0.17 |
ENSRNOT00000086468
|
Ramp3
|
receptor activity modifying protein 3 |
| chr8_-_68525911 | 0.17 |
ENSRNOT00000080588
ENSRNOT00000011109 |
Iqch
|
IQ motif containing H |
| chr15_+_34352914 | 0.17 |
ENSRNOT00000067150
|
Gmpr2
|
guanosine monophosphate reductase 2 |
| chr1_-_196884302 | 0.17 |
ENSRNOT00000089464
|
Nsmce1
|
NSE1 homolog, SMC5-SMC6 complex component |
| chr13_-_77959089 | 0.16 |
ENSRNOT00000003586
|
Cacybp
|
calcyclin binding protein |
| chr5_-_172488822 | 0.16 |
ENSRNOT00000019620
|
Rer1
|
retention in endoplasmic reticulum sorting receptor 1 |
| chrX_+_32495809 | 0.16 |
ENSRNOT00000020999
|
RGD1565844
|
similar to RIKEN cDNA 1700045I19 |
| chr14_+_85113578 | 0.16 |
ENSRNOT00000011158
|
Nipsnap1
|
nipsnap homolog 1 |
| chr3_-_148313810 | 0.16 |
ENSRNOT00000010762
|
Bcl2l1
|
Bcl2-like 1 |
| chr8_-_115388266 | 0.16 |
ENSRNOT00000018122
|
Tex264
|
testis expressed 264 |
| chr19_+_39357791 | 0.16 |
ENSRNOT00000086435
ENSRNOT00000015006 |
Cyb5b
|
cytochrome b5 type B |
| chr14_-_106393670 | 0.16 |
ENSRNOT00000011429
|
Mdh1
|
malate dehydrogenase 1 |
| chrX_-_79909678 | 0.16 |
ENSRNOT00000050336
|
Brwd3
|
bromodomain and WD repeat domain containing 3 |
| chr19_+_26142720 | 0.16 |
ENSRNOT00000005270
|
RGD1564093
|
similar to RIKEN cDNA 2310036O22 |
| chr9_-_94601852 | 0.15 |
ENSRNOT00000022485
|
Ngef
|
neuronal guanine nucleotide exchange factor |
| chr7_+_54859326 | 0.15 |
ENSRNOT00000039141
|
Caps2
|
calcyphosine 2 |
| chr9_-_64096265 | 0.15 |
ENSRNOT00000013502
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr1_-_267598639 | 0.15 |
ENSRNOT00000031925
|
Cfap43
|
cilia and flagella associated protein 43 |
| chr5_+_164808323 | 0.15 |
ENSRNOT00000011005
|
Nppa
|
natriuretic peptide A |
| chr3_-_58181971 | 0.15 |
ENSRNOT00000002076
|
Dlx2
|
distal-less homeobox 2 |
| chr6_-_135259603 | 0.15 |
ENSRNOT00000010529
|
Mok
|
MOK protein kinase |
| chr18_+_63394900 | 0.15 |
ENSRNOT00000024126
|
Psmg2
|
proteasome assembly chaperone 2 |
| chr1_+_170242846 | 0.15 |
ENSRNOT00000023751
|
Cnga4
|
cyclic nucleotide gated channel alpha 4 |
| chr3_+_153491151 | 0.15 |
ENSRNOT00000047047
|
Manbal
|
mannosidase beta like |
| chr10_-_19715832 | 0.15 |
ENSRNOT00000040278
|
RGD1564698
|
similar to ribosomal protein S10 |
| chr7_-_126028279 | 0.15 |
ENSRNOT00000044883
|
Smc1b
|
structural maintenance of chromosomes 1B |
| chr7_+_117420788 | 0.15 |
ENSRNOT00000049198
|
Wdr97
|
WD repeat domain 97 |
| chr8_-_58542844 | 0.14 |
ENSRNOT00000012041
|
Elmod1
|
ELMO domain containing 1 |
| chr19_+_37795658 | 0.14 |
ENSRNOT00000025686
|
Tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr3_+_7051695 | 0.14 |
ENSRNOT00000013548
|
Mrps2
|
mitochondrial ribosomal protein S2 |
| chr4_-_10020022 | 0.14 |
ENSRNOT00000044020
|
Armc10
|
armadillo repeat containing 10 |
| chr3_-_10371240 | 0.14 |
ENSRNOT00000012075
|
Ass1
|
argininosuccinate synthase 1 |
| chr14_+_2325308 | 0.14 |
ENSRNOT00000000072
|
Atp5i
|
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit E |
| chr1_+_86949413 | 0.14 |
ENSRNOT00000064153
|
Sirt2
|
sirtuin 2 |
| chr1_-_225631468 | 0.14 |
ENSRNOT00000072579
|
AABR07006232.1
|
|
| chr11_-_83483513 | 0.14 |
ENSRNOT00000084380
|
AABR07034673.1
|
|
| chr8_-_87315955 | 0.14 |
ENSRNOT00000081437
|
Filip1
|
filamin A interacting protein 1 |
| chr3_+_56802714 | 0.14 |
ENSRNOT00000077551
|
Erich2
|
glutamate-rich 2 |
| chr8_+_64364741 | 0.14 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr6_-_10899200 | 0.13 |
ENSRNOT00000089104
|
Mcfd2
|
multiple coagulation factor deficiency 2 |
| chr10_-_65424802 | 0.13 |
ENSRNOT00000018468
|
Traf4
|
Tnf receptor associated factor 4 |
| chr11_-_90234286 | 0.13 |
ENSRNOT00000071986
|
Efcab1
|
EF hand calcium binding domain 1 |
| chr1_-_142720257 | 0.13 |
ENSRNOT00000032294
|
Wdr73
|
WD repeat domain 73 |
| chr4_+_21317695 | 0.13 |
ENSRNOT00000007572
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr20_-_32296840 | 0.13 |
ENSRNOT00000080727
|
Stox1
|
storkhead box 1 |
| chr1_+_265298868 | 0.13 |
ENSRNOT00000023278
|
Dpcd
|
deleted in primary ciliary dyskinesia |
| chr1_-_142020525 | 0.13 |
ENSRNOT00000042558
|
Cib1
|
calcium and integrin binding 1 |
| chr6_+_111223026 | 0.13 |
ENSRNOT00000074634
|
Samd15
|
sterile alpha motif domain containing 15 |
| chrX_+_20351486 | 0.13 |
ENSRNOT00000093675
ENSRNOT00000047444 |
Wnk3
|
WNK lysine deficient protein kinase 3 |
| chr1_-_64021321 | 0.13 |
ENSRNOT00000090819
|
Rps9
|
ribosomal protein S9 |
| chr4_+_25435873 | 0.13 |
ENSRNOT00000000018
|
Steap1
|
STEAP family member 1 |
| chr2_+_211050360 | 0.13 |
ENSRNOT00000026928
|
Psma5
|
proteasome subunit alpha 5 |
| chr16_-_39970532 | 0.13 |
ENSRNOT00000071331
|
Spata4
|
spermatogenesis associated 4 |
| chrX_+_122808605 | 0.13 |
ENSRNOT00000017567
|
Zcchc12
|
zinc finger CCHC-type containing 12 |
| chr7_+_120140460 | 0.12 |
ENSRNOT00000040513
ENSRNOT00000073905 |
Pdxp
|
pyridoxal phosphatase |
| chr11_-_31892531 | 0.12 |
ENSRNOT00000033745
|
Cryzl1
|
crystallin zeta like 1 |
| chr9_+_81644355 | 0.12 |
ENSRNOT00000071700
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr12_+_47024442 | 0.12 |
ENSRNOT00000001545
|
Cox6a1
|
cytochrome c oxidase subunit 6A1 |
| chr3_+_148438939 | 0.12 |
ENSRNOT00000064196
|
Ttll9
|
tubulin tyrosine ligase like 9 |
| chr20_-_32353677 | 0.12 |
ENSRNOT00000035355
|
Stox1
|
storkhead box 1 |
| chr8_-_63291966 | 0.12 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr16_-_7758189 | 0.12 |
ENSRNOT00000026588
|
Hacl1
|
2-hydroxyacyl-CoA lyase 1 |
| chr10_-_110585376 | 0.12 |
ENSRNOT00000054917
|
Rab40b
|
Rab40b, member RAS oncogene family |
| chr16_+_51748970 | 0.12 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chr3_-_56030744 | 0.12 |
ENSRNOT00000089637
|
Ccdc173
|
coiled-coil domain containing 173 |
| chrX_-_80354604 | 0.12 |
ENSRNOT00000049790
|
RGD1562039
|
similar to MGC82337 protein |
| chr11_-_61470046 | 0.12 |
ENSRNOT00000073436
|
LOC102553099
|
N-alpha-acetyltransferase 50-like |
| chr1_-_229601032 | 0.12 |
ENSRNOT00000016690
|
Cntf
|
ciliary neurotrophic factor |
| chr12_+_23954574 | 0.11 |
ENSRNOT00000046343
|
Styxl1
|
serine/threonine/tyrosine interacting-like 1 |
| chr1_+_78739930 | 0.11 |
ENSRNOT00000021976
|
Strn4
|
striatin 4 |
| chrX_-_115073890 | 0.11 |
ENSRNOT00000006638
|
Capn6
|
calpain 6 |
| chr2_-_250600517 | 0.11 |
ENSRNOT00000016872
|
Hs2st1
|
heparan sulfate 2-O-sulfotransferase 1 |
| chr12_-_20660304 | 0.11 |
ENSRNOT00000091323
|
RGD1561730
|
similar to cell surface receptor FDFACT |
| chr4_-_66380734 | 0.11 |
ENSRNOT00000009194
|
Klrg2
|
killer cell lectin like receptor G2 |
| chr6_-_94932806 | 0.11 |
ENSRNOT00000006346
|
Ccdc175
|
coiled-coil domain containing 175 |
| chr8_+_117221367 | 0.11 |
ENSRNOT00000073445
|
LOC680045
|
hypothetical protein LOC680045 |
| chr10_-_11878792 | 0.11 |
ENSRNOT00000064964
|
Cluap1
|
clusterin associated protein 1 |
| chr10_-_38782419 | 0.11 |
ENSRNOT00000073964
|
Uqcrq
|
ubiquinol-cytochrome c reductase, complex III subunit VII |
| chr16_-_7406951 | 0.11 |
ENSRNOT00000066508
|
Dnah1
|
dynein, axonemal, heavy chain 1 |
| chr10_-_34961349 | 0.11 |
ENSRNOT00000004885
|
LOC103689931
|
heterogeneous nuclear ribonucleoprotein A/B |
| chr5_+_113592919 | 0.11 |
ENSRNOT00000011336
|
Ift74
|
intraflagellar transport 74 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rfx2_Rfx7
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.2 | 0.8 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.1 | 0.4 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.1 | 0.2 | GO:1900195 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.1 | 0.2 | GO:0016476 | regulation of embryonic cell shape(GO:0016476) |
| 0.1 | 0.3 | GO:0071883 | activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.1 | 0.2 | GO:0002432 | granuloma formation(GO:0002432) T-helper 1 cell activation(GO:0035711) |
| 0.1 | 0.3 | GO:1990564 | protein polyufmylation(GO:1990564) protein K69-linked ufmylation(GO:1990592) |
| 0.1 | 0.9 | GO:0042407 | cristae formation(GO:0042407) |
| 0.1 | 0.4 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.1 | 0.2 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.1 | 0.2 | GO:1902304 | positive regulation of potassium ion export(GO:1902304) |
| 0.1 | 0.2 | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.1 | GO:0000053 | argininosuccinate metabolic process(GO:0000053) |
| 0.0 | 0.8 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.2 | GO:0045903 | positive regulation of translational fidelity(GO:0045903) |
| 0.0 | 0.6 | GO:1903830 | magnesium ion transport(GO:0015693) magnesium ion transmembrane transport(GO:1903830) |
| 0.0 | 0.4 | GO:0036159 | inner dynein arm assembly(GO:0036159) |
| 0.0 | 0.3 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.1 | GO:0021678 | third ventricle development(GO:0021678) |
| 0.0 | 0.1 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.1 | GO:2000682 | positive regulation of rubidium ion transport(GO:2000682) positive regulation of rubidium ion transmembrane transporter activity(GO:2000688) |
| 0.0 | 0.3 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.1 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.2 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.6 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.1 | GO:0033505 | floor plate formation(GO:0021508) floor plate morphogenesis(GO:0033505) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0021539 | subthalamus development(GO:0021539) |
| 0.0 | 0.3 | GO:0039536 | negative regulation of RIG-I signaling pathway(GO:0039536) |
| 0.0 | 0.5 | GO:0003351 | epithelial cilium movement(GO:0003351) |
| 0.0 | 0.2 | GO:0034486 | vacuolar transmembrane transport(GO:0034486) chaperone-mediated protein transport involved in chaperone-mediated autophagy(GO:0061741) |
| 0.0 | 0.2 | GO:1905232 | cellular response to L-glutamate(GO:1905232) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:1903002 | regulation of lipid transport across blood brain barrier(GO:1903000) positive regulation of lipid transport across blood brain barrier(GO:1903002) |
| 0.0 | 0.1 | GO:1902953 | endoplasmic reticulum membrane organization(GO:0090158) positive regulation of ER to Golgi vesicle-mediated transport(GO:1902953) |
| 0.0 | 0.1 | GO:0048691 | positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) |
| 0.0 | 0.5 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0006348 | chromatin silencing at telomere(GO:0006348) |
| 0.0 | 0.3 | GO:0002318 | myeloid progenitor cell differentiation(GO:0002318) |
| 0.0 | 0.1 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:2001013 | positive regulation of macrophage apoptotic process(GO:2000111) epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.2 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.1 | GO:0009182 | purine deoxyribonucleotide biosynthetic process(GO:0009153) purine deoxyribonucleoside diphosphate metabolic process(GO:0009182) dGDP metabolic process(GO:0046066) |
| 0.0 | 0.1 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.1 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.1 | GO:0034080 | CENP-A containing nucleosome assembly(GO:0034080) CENP-A containing chromatin organization(GO:0061641) |
| 0.0 | 0.1 | GO:1904569 | response to selenite ion(GO:0072714) regulation of selenocysteine incorporation(GO:1904569) |
| 0.0 | 0.1 | GO:0070900 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.2 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0001561 | fatty acid alpha-oxidation(GO:0001561) |
| 0.0 | 0.2 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0070946 | neutrophil mediated killing of gram-positive bacterium(GO:0070946) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.1 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) |
| 0.0 | 0.3 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:1902774 | terminal button organization(GO:0072553) late endosome to lysosome transport(GO:1902774) |
| 0.0 | 0.1 | GO:0071899 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0097461 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.0 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.0 | 0.1 | GO:1903751 | regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903750) negative regulation of intrinsic apoptotic signaling pathway in response to hydrogen peroxide(GO:1903751) |
| 0.0 | 0.2 | GO:0086103 | G-protein coupled receptor signaling pathway involved in heart process(GO:0086103) |
| 0.0 | 0.1 | GO:0045048 | protein insertion into ER membrane(GO:0045048) |
| 0.0 | 0.0 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.0 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.1 | GO:0002176 | male germ cell proliferation(GO:0002176) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.0 | 1.1 | GO:1903955 | positive regulation of protein targeting to mitochondrion(GO:1903955) |
| 0.0 | 0.2 | GO:0045653 | negative regulation of megakaryocyte differentiation(GO:0045653) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.1 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) |
| 0.0 | 0.1 | GO:0009134 | nucleoside diphosphate catabolic process(GO:0009134) |
| 0.0 | 0.2 | GO:0071340 | skeletal muscle acetylcholine-gated channel clustering(GO:0071340) |
| 0.0 | 0.1 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.0 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0018243 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:0003341 | cilium movement(GO:0003341) |
| 0.0 | 0.1 | GO:0098734 | macromolecule depalmitoylation(GO:0098734) |
| 0.0 | 0.1 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.1 | GO:1902951 | negative regulation of dendritic spine maintenance(GO:1902951) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0061617 | MICOS complex(GO:0061617) |
| 0.1 | 0.6 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.1 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.1 | 0.2 | GO:0046691 | intracellular canaliculus(GO:0046691) |
| 0.1 | 0.3 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.2 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.1 | 0.2 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.2 | GO:0036156 | inner dynein arm(GO:0036156) |
| 0.0 | 0.2 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
| 0.0 | 1.2 | GO:0097228 | sperm principal piece(GO:0097228) |
| 0.0 | 0.1 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 0.6 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.3 | GO:0017071 | intracellular cyclic nucleotide activated cation channel complex(GO:0017071) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.1 | GO:0071942 | XPC complex(GO:0071942) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.2 | GO:0005818 | astral microtubule(GO:0000235) aster(GO:0005818) |
| 0.0 | 0.1 | GO:0034991 | nuclear meiotic cohesin complex(GO:0034991) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 1.1 | GO:0036126 | sperm flagellum(GO:0036126) |
| 0.0 | 0.3 | GO:0019013 | viral nucleocapsid(GO:0019013) viral capsid(GO:0019028) |
| 0.0 | 0.0 | GO:0097451 | glial limiting end-foot(GO:0097451) |
| 0.0 | 0.1 | GO:0044447 | axoneme part(GO:0044447) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.1 | GO:0005750 | mitochondrial respiratory chain complex III(GO:0005750) respiratory chain complex III(GO:0045275) |
| 0.0 | 0.1 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.2 | GO:0048188 | Set1C/COMPASS complex(GO:0048188) |
| 0.0 | 0.2 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.1 | 0.2 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.1 | 0.3 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.1 | 0.2 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.1 | 0.4 | GO:0005432 | calcium:sodium antiporter activity(GO:0005432) |
| 0.1 | 0.2 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.1 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.2 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.6 | GO:0004576 | oligosaccharyl transferase activity(GO:0004576) dolichyl-diphosphooligosaccharide-protein glycotransferase activity(GO:0004579) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:1990932 | 5.8S rRNA binding(GO:1990932) |
| 0.0 | 0.3 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.1 | GO:0008427 | calcium-dependent protein kinase inhibitor activity(GO:0008427) |
| 0.0 | 0.1 | GO:0046911 | metal chelating activity(GO:0046911) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.3 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.4 | GO:0030550 | acetylcholine receptor inhibitor activity(GO:0030550) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0004945 | angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.3 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.3 | GO:0001056 | RNA polymerase III activity(GO:0001056) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.2 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 0.3 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0008121 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 1.1 | GO:0004843 | thiol-dependent ubiquitin-specific protease activity(GO:0004843) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0070739 | protein-glutamic acid ligase activity(GO:0070739) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.1 | GO:0015252 | hydrogen ion channel activity(GO:0015252) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.1 | GO:1901611 | phosphatidylglycerol binding(GO:1901611) cardiolipin binding(GO:1901612) |
| 0.0 | 0.1 | GO:0070016 | armadillo repeat domain binding(GO:0070016) |
| 0.0 | 0.0 | GO:0016672 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0047134 | protein-disulfide reductase activity(GO:0047134) |
| 0.0 | 0.0 | GO:0003854 | 3-beta-hydroxy-delta5-steroid dehydrogenase activity(GO:0003854) |
| 0.0 | 0.1 | GO:0019869 | chloride channel inhibitor activity(GO:0019869) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.1 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.3 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.3 | PID ARF 3PATHWAY | Arf1 pathway |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.3 | REACTOME RNA POL III CHAIN ELONGATION | Genes involved in RNA Polymerase III Chain Elongation |
| 0.0 | 0.2 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.1 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME GENERATION OF SECOND MESSENGER MOLECULES | Genes involved in Generation of second messenger molecules |