Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
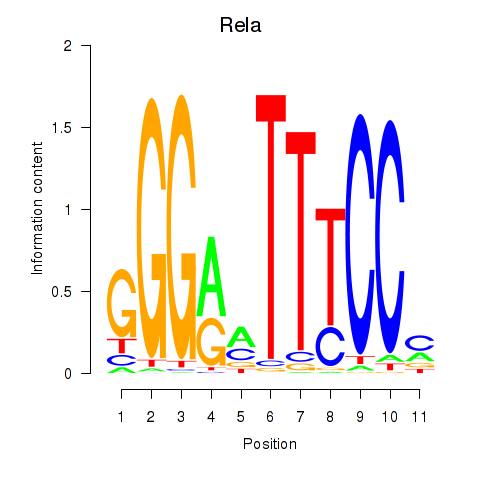
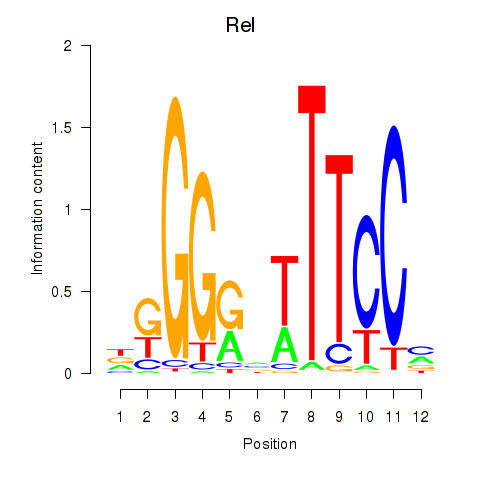
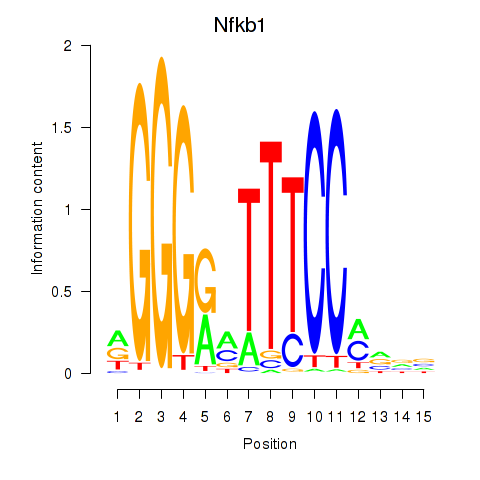
Results for Rela_Rel_Nfkb1
Z-value: 0.62
Transcription factors associated with Rela_Rel_Nfkb1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rela
|
ENSRNOG00000030888 | RELA proto-oncogene, NF-kB subunit |
|
Rel
|
ENSRNOG00000054437 | REL proto-oncogene, NF-kB subunit |
|
Nfkb1
|
ENSRNOG00000023258 | nuclear factor kappa B subunit 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Nfkb1 | rn6_v1_chr2_-_240866689_240866689 | -0.73 | 1.6e-01 | Click! |
| Rel | rn6_v1_chr14_-_108509892_108509892 | -0.47 | 4.2e-01 | Click! |
| Rela | rn6_v1_chr1_+_220992770_220992770 | -0.23 | 7.1e-01 | Click! |
Activity profile of Rela_Rel_Nfkb1 motif
Sorted Z-values of Rela_Rel_Nfkb1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_76270457 | 1.03 |
ENSRNOT00000009894
|
Nfkbia
|
NFKB inhibitor alpha |
| chr14_-_18745457 | 0.43 |
ENSRNOT00000003778
|
Cxcl1
|
C-X-C motif chemokine ligand 1 |
| chr2_+_189955836 | 0.35 |
ENSRNOT00000078351
|
S100a3
|
S100 calcium binding protein A3 |
| chr17_+_76002275 | 0.33 |
ENSRNOT00000092665
ENSRNOT00000086701 |
Echdc3
|
enoyl CoA hydratase domain containing 3 |
| chr10_-_85947938 | 0.29 |
ENSRNOT00000037318
ENSRNOT00000082427 |
Arl5c
|
ADP-ribosylation factor like GTPase 5C |
| chr1_+_163445527 | 0.27 |
ENSRNOT00000020520
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr20_-_4921348 | 0.25 |
ENSRNOT00000082497
ENSRNOT00000041151 |
RT1-CE4
|
RT1 class I, locus CE4 |
| chr18_-_28516209 | 0.25 |
ENSRNOT00000072580
|
Ecscr
|
endothelial cell surface expressed chemotaxis and apoptosis regulator |
| chr9_-_11114843 | 0.24 |
ENSRNOT00000072031
|
Ebi3
|
Epstein-Barr virus induced 3 |
| chr11_+_47243342 | 0.22 |
ENSRNOT00000041116
|
Nfkbiz
|
NFKB inhibitor zeta |
| chr20_+_5184515 | 0.21 |
ENSRNOT00000089411
|
LOC103694381
|
lymphotoxin-beta |
| chr20_-_4863011 | 0.21 |
ENSRNOT00000079503
|
Ltb
|
lymphotoxin beta |
| chr5_-_169017295 | 0.20 |
ENSRNOT00000067481
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr5_-_153924896 | 0.20 |
ENSRNOT00000065247
|
Grhl3
|
grainyhead-like transcription factor 3 |
| chr3_-_11417546 | 0.19 |
ENSRNOT00000018776
|
Lcn2
|
lipocalin 2 |
| chr7_-_11648322 | 0.19 |
ENSRNOT00000026871
|
Gadd45b
|
growth arrest and DNA-damage-inducible, beta |
| chr9_+_9842585 | 0.18 |
ENSRNOT00000070806
|
Cd70
|
Cd70 molecule |
| chr4_+_171250818 | 0.17 |
ENSRNOT00000040576
|
Ptpro
|
protein tyrosine phosphatase, receptor type, O |
| chr1_-_80744831 | 0.17 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chr17_-_13393243 | 0.17 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr14_-_18733391 | 0.16 |
ENSRNOT00000003745
|
Cxcl2
|
C-X-C motif chemokine ligand 2 |
| chr7_-_28711761 | 0.16 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr4_-_157266018 | 0.16 |
ENSRNOT00000019570
|
LOC100911713
|
protein C10-like |
| chr17_+_56109549 | 0.16 |
ENSRNOT00000022190
|
Map3k8
|
mitogen-activated protein kinase kinase kinase 8 |
| chr5_+_135562034 | 0.15 |
ENSRNOT00000056967
|
Ccdc163
|
coiled-coil domain containing 163 |
| chr20_-_3439983 | 0.15 |
ENSRNOT00000080822
ENSRNOT00000001099 |
Ier3
|
immediate early response 3 |
| chr5_-_167999853 | 0.15 |
ENSRNOT00000087402
|
Park7
|
Parkinsonism associated deglycase |
| chr3_+_2643610 | 0.14 |
ENSRNOT00000037169
|
Fut7
|
fucosyltransferase 7 |
| chr7_+_54247460 | 0.14 |
ENSRNOT00000005361
|
Phlda1
|
pleckstrin homology-like domain, family A, member 1 |
| chr1_+_80135391 | 0.14 |
ENSRNOT00000021893
|
Gpr4
|
G protein-coupled receptor 4 |
| chr20_-_4863198 | 0.14 |
ENSRNOT00000001108
|
Ltb
|
lymphotoxin beta |
| chr1_+_86948918 | 0.14 |
ENSRNOT00000084839
|
Sirt2
|
sirtuin 2 |
| chr19_-_55257876 | 0.13 |
ENSRNOT00000017564
|
Cyba
|
cytochrome b-245 alpha chain |
| chr18_-_15540177 | 0.13 |
ENSRNOT00000022113
|
Ttr
|
transthyretin |
| chr11_+_33909439 | 0.13 |
ENSRNOT00000002310
|
Cbr3
|
carbonyl reductase 3 |
| chr9_+_20279938 | 0.13 |
ENSRNOT00000075612
|
Grcc10
|
gene rich cluster, C10 gene |
| chr19_+_561727 | 0.13 |
ENSRNOT00000016259
ENSRNOT00000081547 |
Rrad
|
RRAD, Ras related glycolysis inhibitor and calcium channel regulator |
| chr8_+_133029625 | 0.13 |
ENSRNOT00000008809
|
Ccr3
|
C-C motif chemokine receptor 3 |
| chr10_-_13081136 | 0.12 |
ENSRNOT00000005718
|
Kremen2
|
kringle containing transmembrane protein 2 |
| chr5_+_168019058 | 0.12 |
ENSRNOT00000077202
|
Tnfrsf9
|
TNF receptor superfamily member 9 |
| chr2_+_195603599 | 0.12 |
ENSRNOT00000028263
|
C2cd4d
|
C2 calcium-dependent domain containing 4D |
| chr13_-_36022197 | 0.12 |
ENSRNOT00000091280
|
Cfap221
|
cilia and flagella associated protein 221 |
| chr4_-_28953067 | 0.11 |
ENSRNOT00000013989
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr1_-_86948845 | 0.11 |
ENSRNOT00000027212
|
Nfkbib
|
NFKB inhibitor beta |
| chrX_-_32355296 | 0.11 |
ENSRNOT00000081652
ENSRNOT00000065075 |
Ap1s2
|
adaptor-related protein complex 1, sigma 2 subunit |
| chr1_+_211582077 | 0.11 |
ENSRNOT00000023619
|
Pwwp2b
|
PWWP domain containing 2B |
| chr1_-_259357056 | 0.11 |
ENSRNOT00000022000
|
Pdlim1
|
PDZ and LIM domain 1 |
| chr10_-_97582188 | 0.11 |
ENSRNOT00000005076
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr15_+_50891127 | 0.11 |
ENSRNOT00000020728
|
Stc1
|
stanniocalcin 1 |
| chr1_+_82473737 | 0.11 |
ENSRNOT00000028029
|
B9d2
|
B9 protein domain 2 |
| chr10_+_110139783 | 0.11 |
ENSRNOT00000054939
|
Slc16a3
|
solute carrier family 16 member 3 |
| chr19_+_37229120 | 0.11 |
ENSRNOT00000076335
|
Hsf4
|
heat shock transcription factor 4 |
| chr6_+_135866739 | 0.11 |
ENSRNOT00000013460
|
Exoc3l4
|
exocyst complex component 3-like 4 |
| chr13_-_52088780 | 0.11 |
ENSRNOT00000008754
|
Elf3
|
E74-like factor 3 |
| chr8_-_48850671 | 0.11 |
ENSRNOT00000016580
|
Cxcr5
|
C-X-C motif chemokine receptor 5 |
| chr13_-_90814119 | 0.10 |
ENSRNOT00000011208
|
Tagln2
|
transgelin 2 |
| chr5_-_137372993 | 0.10 |
ENSRNOT00000092823
|
Tmem125
|
transmembrane protein 125 |
| chr10_+_86303727 | 0.10 |
ENSRNOT00000037752
|
Ppp1r1b
|
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chr20_+_14095914 | 0.10 |
ENSRNOT00000093404
|
Gucd1
|
guanylyl cyclase domain containing 1 |
| chr14_+_17195014 | 0.10 |
ENSRNOT00000031667
|
Cxcl11
|
C-X-C motif chemokine ligand 11 |
| chr8_+_23113048 | 0.10 |
ENSRNOT00000029577
|
Cnn1
|
calponin 1 |
| chr10_+_14240219 | 0.10 |
ENSRNOT00000020233
|
Igfals
|
insulin-like growth factor binding protein, acid labile subunit |
| chr14_-_34218961 | 0.09 |
ENSRNOT00000072588
|
LOC686911
|
similar to Exocyst complex component 1 (Exocyst complex component Sec3) |
| chr5_+_135029955 | 0.09 |
ENSRNOT00000074860
|
LOC100911669
|
uncharacterized LOC100911669 |
| chr18_+_56071478 | 0.09 |
ENSRNOT00000025344
ENSRNOT00000025354 |
Cd74
|
CD74 molecule |
| chr20_-_3299580 | 0.09 |
ENSRNOT00000050373
|
Gnl1
|
G protein nucleolar 1 |
| chr1_+_202432366 | 0.09 |
ENSRNOT00000027681
|
Plpp4
|
phospholipid phosphatase 4 |
| chr7_-_28932641 | 0.09 |
ENSRNOT00000059487
|
Dram1
|
DNA-damage regulated autophagy modulator 1 |
| chr11_+_31694339 | 0.09 |
ENSRNOT00000002779
|
Ifngr2
|
interferon gamma receptor 2 |
| chr14_-_18839595 | 0.09 |
ENSRNOT00000078746
|
Cxcl3
|
chemokine (C-X-C motif) ligand 3 |
| chr5_-_147584038 | 0.08 |
ENSRNOT00000010983
|
Zbtb8a
|
zinc finger and BTB domain containing 8a |
| chrX_+_74497262 | 0.08 |
ENSRNOT00000003899
|
Zcchc13
|
zinc finger CCHC-type containing 13 |
| chr1_-_167347662 | 0.08 |
ENSRNOT00000027641
ENSRNOT00000076592 |
Rhog
|
ras homolog family member G |
| chr10_-_10767389 | 0.08 |
ENSRNOT00000066754
|
Smim22
|
small integral membrane protein 22 |
| chr5_-_166924136 | 0.08 |
ENSRNOT00000085251
|
Spsb1
|
splA/ryanodine receptor domain and SOCS box containing 1 |
| chr19_-_6031655 | 0.08 |
ENSRNOT00000092016
|
AABR07042733.2
|
|
| chr4_-_93406182 | 0.08 |
ENSRNOT00000007437
|
AABR07060778.1
|
|
| chr4_-_30338679 | 0.08 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr1_+_225068009 | 0.08 |
ENSRNOT00000026651
|
Ubxn1
|
UBX domain protein 1 |
| chr4_+_83713666 | 0.08 |
ENSRNOT00000086473
|
Creb5
|
cAMP responsive element binding protein 5 |
| chr5_+_98469047 | 0.08 |
ENSRNOT00000041374
|
Lurap1l
|
leucine rich adaptor protein 1-like |
| chr3_+_113415774 | 0.08 |
ENSRNOT00000056151
|
Serf2
|
small EDRK-rich factor 2 |
| chr9_+_81644975 | 0.08 |
ENSRNOT00000057480
|
Catip
|
ciliogenesis associated TTC17 interacting protein |
| chr1_+_84470829 | 0.08 |
ENSRNOT00000025472
|
Ttc9b
|
tetratricopeptide repeat domain 9B |
| chr9_+_88918433 | 0.07 |
ENSRNOT00000021730
|
Ccl20
|
C-C motif chemokine ligand 20 |
| chr10_+_77537340 | 0.07 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr6_+_135890931 | 0.07 |
ENSRNOT00000042973
|
Tnfaip2
|
TNF alpha induced protein 2 |
| chr5_-_168000083 | 0.07 |
ENSRNOT00000091206
|
Park7
|
Parkinsonism associated deglycase |
| chr3_-_9738752 | 0.07 |
ENSRNOT00000045993
|
Ptges
|
prostaglandin E synthase |
| chr4_+_78458625 | 0.07 |
ENSRNOT00000049891
|
Tmem176a
|
transmembrane protein 176A |
| chr2_+_154604832 | 0.07 |
ENSRNOT00000013777
|
Vom2r44
|
vomeronasal 2 receptor 44 |
| chr7_-_145154131 | 0.07 |
ENSRNOT00000055271
|
Ppp1r1a
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chrX_-_15761932 | 0.07 |
ENSRNOT00000015641
ENSRNOT00000091146 |
Foxp3
|
forkhead box P3 |
| chr1_+_177569618 | 0.07 |
ENSRNOT00000090042
|
Tead1
|
TEA domain transcription factor 1 |
| chr8_-_6076598 | 0.07 |
ENSRNOT00000090774
|
Birc3
|
baculoviral IAP repeat-containing 3 |
| chr17_+_14469488 | 0.07 |
ENSRNOT00000060670
|
AABR07027111.1
|
|
| chr17_-_1999596 | 0.07 |
ENSRNOT00000072220
|
1190003K10Rik
|
RIKEN cDNA 1190003K10 gene |
| chr1_-_215553451 | 0.07 |
ENSRNOT00000027407
|
Ctsd
|
cathepsin D |
| chr10_-_70744315 | 0.07 |
ENSRNOT00000014865
|
Ccl5
|
C-C motif chemokine ligand 5 |
| chr4_-_157439507 | 0.07 |
ENSRNOT00000081802
|
Ptms
|
parathymosin |
| chr10_-_88172910 | 0.07 |
ENSRNOT00000046956
|
Krt42
|
keratin 42 |
| chr4_+_78080310 | 0.07 |
ENSRNOT00000035906
|
Sspo
|
SCO-spondin |
| chr19_-_37912027 | 0.07 |
ENSRNOT00000026462
|
Psmb10
|
proteasome subunit beta 10 |
| chr4_+_44321883 | 0.06 |
ENSRNOT00000091095
|
Tes
|
testin LIM domain protein |
| chr4_+_145399913 | 0.06 |
ENSRNOT00000012177
|
Jagn1
|
jagunal homolog 1 |
| chr7_+_116632506 | 0.06 |
ENSRNOT00000009811
|
Gpihbp1
|
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr1_-_226657408 | 0.06 |
ENSRNOT00000028102
|
Tkfc
|
triokinase and FMN cyclase |
| chr18_+_81694808 | 0.06 |
ENSRNOT00000020446
|
Cyb5a
|
cytochrome b5 type A |
| chr2_-_204254699 | 0.06 |
ENSRNOT00000021487
|
Mab21l3
|
mab-21 like 3 |
| chr6_+_28382962 | 0.06 |
ENSRNOT00000016976
|
Pomc
|
proopiomelanocortin |
| chr20_-_3298835 | 0.06 |
ENSRNOT00000083617
|
Gnl1
|
G protein nucleolar 1 |
| chr1_-_24190896 | 0.06 |
ENSRNOT00000016121
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr17_+_80882666 | 0.06 |
ENSRNOT00000024430
|
Vim
|
vimentin |
| chr1_-_3849080 | 0.06 |
ENSRNOT00000045301
|
RGD1560633
|
similar to ribosomal protein L27a |
| chr9_+_82647071 | 0.06 |
ENSRNOT00000027135
|
Asic4
|
acid sensing ion channel subunit family member 4 |
| chr12_+_27155587 | 0.06 |
ENSRNOT00000044800
|
AABR07035916.1
|
|
| chr4_-_78458179 | 0.06 |
ENSRNOT00000078473
ENSRNOT00000011327 |
Tmem176b
|
transmembrane protein 176B |
| chr4_-_157252104 | 0.06 |
ENSRNOT00000082739
|
Ptpn6
|
protein tyrosine phosphatase, non-receptor type 6 |
| chr7_-_71048383 | 0.06 |
ENSRNOT00000005693
|
Gpr182
|
G protein-coupled receptor 182 |
| chr20_+_4966817 | 0.06 |
ENSRNOT00000081527
ENSRNOT00000081265 |
Lsm2
|
LSM2 homolog, U6 small nuclear RNA and mRNA degradation associated |
| chr1_+_41325462 | 0.06 |
ENSRNOT00000081017
ENSRNOT00000078494 ENSRNOT00000088168 |
Esr1
|
estrogen receptor 1 |
| chr10_-_95934345 | 0.06 |
ENSRNOT00000004349
|
Cacng1
|
calcium voltage-gated channel auxiliary subunit gamma 1 |
| chr9_+_82053581 | 0.06 |
ENSRNOT00000086375
|
Wnt10a
|
wingless-type MMTV integration site family, member 10A |
| chr1_+_240776969 | 0.06 |
ENSRNOT00000049732
|
RGD1561453
|
similar to ribosomal protein S12 |
| chr3_-_175479034 | 0.06 |
ENSRNOT00000086761
|
Hrh3
|
histamine receptor H3 |
| chr10_-_87407634 | 0.06 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr1_-_85517360 | 0.06 |
ENSRNOT00000026114
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr14_+_4362717 | 0.06 |
ENSRNOT00000002887
|
Barhl2
|
BarH-like homeobox 2 |
| chr5_-_109651730 | 0.06 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr7_-_144993652 | 0.06 |
ENSRNOT00000087748
|
Itga5
|
integrin subunit alpha 5 |
| chr16_+_61954809 | 0.05 |
ENSRNOT00000068011
|
Rbpms
|
RNA binding protein with multiple splicing |
| chr18_+_60745035 | 0.05 |
ENSRNOT00000045098
|
LOC502176
|
hypothetical protein LOC502176 |
| chr20_-_3978845 | 0.05 |
ENSRNOT00000000532
|
Psmb9
|
proteasome subunit beta 9 |
| chr15_-_25505691 | 0.05 |
ENSRNOT00000074488
|
LOC100911492
|
homeobox protein OTX2-like |
| chr1_-_213811901 | 0.05 |
ENSRNOT00000020265
|
Ifitm3
|
interferon induced transmembrane protein 3 |
| chr3_-_142752325 | 0.05 |
ENSRNOT00000006200
|
Thbd
|
thrombomodulin |
| chr13_-_77959089 | 0.05 |
ENSRNOT00000003586
|
Cacybp
|
calcyclin binding protein |
| chr5_+_156026911 | 0.05 |
ENSRNOT00000046802
|
Rap1gap
|
Rap1 GTPase-activating protein |
| chr13_-_105684374 | 0.05 |
ENSRNOT00000073142
|
Spata17
|
spermatogenesis associated 17 |
| chr10_-_15204143 | 0.05 |
ENSRNOT00000026971
|
Rhbdl1
|
rhomboid like 1 |
| chr1_-_102255459 | 0.05 |
ENSRNOT00000067392
|
Ush1c
|
USH1 protein network component harmonin |
| chr7_-_140356209 | 0.05 |
ENSRNOT00000077856
|
Rnd1
|
Rho family GTPase 1 |
| chr1_-_215536980 | 0.05 |
ENSRNOT00000027344
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chr1_-_129780356 | 0.05 |
ENSRNOT00000077479
|
Arrdc4
|
arrestin domain containing 4 |
| chr7_-_11754508 | 0.05 |
ENSRNOT00000026341
|
Oaz1
|
ornithine decarboxylase antizyme 1 |
| chrX_-_40086870 | 0.05 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr7_-_12918173 | 0.05 |
ENSRNOT00000011010
|
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr15_+_10120206 | 0.05 |
ENSRNOT00000033048
|
Rarb
|
retinoic acid receptor, beta |
| chr12_+_24989298 | 0.05 |
ENSRNOT00000032780
|
Eln
|
elastin |
| chr1_-_215536770 | 0.05 |
ENSRNOT00000078030
|
Ifitm10
|
interferon induced transmembrane protein 10 |
| chrX_-_71121034 | 0.05 |
ENSRNOT00000076590
|
Snx12
|
sorting nexin 12 |
| chr5_-_57267002 | 0.05 |
ENSRNOT00000011455
|
Bag1
|
Bcl2 associated athanogene 1 |
| chr13_-_89433815 | 0.05 |
ENSRNOT00000091541
|
Fcgr2b
|
Fc fragment of IgG receptor IIb |
| chr2_+_60131776 | 0.05 |
ENSRNOT00000080786
|
Prlr
|
prolactin receptor |
| chr4_-_80313411 | 0.05 |
ENSRNOT00000041621
|
RGD1565415
|
similar to ribosomal protein L27a |
| chr15_-_58711872 | 0.05 |
ENSRNOT00000058204
|
Serp2
|
stress-associated endoplasmic reticulum protein family member 2 |
| chr19_+_58505811 | 0.05 |
ENSRNOT00000031717
|
Map10
|
microtubule-associated protein 10 |
| chr14_-_10446909 | 0.05 |
ENSRNOT00000002962
|
Mrps18c
|
mitochondrial ribosomal protein S18C |
| chr7_-_144837395 | 0.05 |
ENSRNOT00000089024
|
Cbx5
|
chromobox 5 |
| chr1_+_184299250 | 0.05 |
ENSRNOT00000030540
|
Insc
|
inscuteable homolog (Drosophila) |
| chr4_+_172119331 | 0.04 |
ENSRNOT00000010579
|
Mgst1
|
microsomal glutathione S-transferase 1 |
| chr10_+_39066716 | 0.04 |
ENSRNOT00000010729
|
Il5
|
interleukin 5 |
| chr4_-_163049084 | 0.04 |
ENSRNOT00000091644
|
Cd69
|
Cd69 molecule |
| chrX_-_104932508 | 0.04 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chr13_-_91228901 | 0.04 |
ENSRNOT00000071728
ENSRNOT00000073643 ENSRNOT00000071897 |
LOC100911825
|
low affinity immunoglobulin gamma Fc region receptor III-like |
| chr2_-_235852708 | 0.04 |
ENSRNOT00000009046
ENSRNOT00000005772 |
Rpl34
|
ribosomal protein L34 |
| chr1_-_266086299 | 0.04 |
ENSRNOT00000026609
|
Cuedc2
|
CUE domain containing 2 |
| chr11_+_72705129 | 0.04 |
ENSRNOT00000073330
|
Apod
|
apolipoprotein D |
| chr3_+_110982553 | 0.04 |
ENSRNOT00000030754
|
LOC691418
|
hypothetical protein LOC691418 |
| chr7_-_12399910 | 0.04 |
ENSRNOT00000021677
|
RGD1562114
|
RGD1562114 |
| chr13_-_50549981 | 0.04 |
ENSRNOT00000003918
ENSRNOT00000080486 |
Golt1a
|
golgi transport 1A |
| chr12_-_22748860 | 0.04 |
ENSRNOT00000001923
|
Cldn15
|
claudin 15 |
| chr20_-_5476193 | 0.04 |
ENSRNOT00000044975
|
Tapbp
|
TAP binding protein |
| chr9_+_42871950 | 0.04 |
ENSRNOT00000089673
|
Arid5a
|
AT-rich interaction domain 5A |
| chr4_-_10517832 | 0.04 |
ENSRNOT00000039953
ENSRNOT00000083964 |
Gsap
|
gamma-secretase activating protein |
| chr9_+_65172194 | 0.04 |
ENSRNOT00000040493
|
AC128084.1
|
|
| chr5_-_143120165 | 0.04 |
ENSRNOT00000012314
|
Zc3h12a
|
zinc finger CCCH type containing 12A |
| chr4_+_71740532 | 0.04 |
ENSRNOT00000023537
|
Zyx
|
zyxin |
| chr20_+_11489456 | 0.04 |
ENSRNOT00000001632
|
Lrrc3
|
leucine rich repeat containing 3 |
| chr1_+_166433109 | 0.04 |
ENSRNOT00000026428
|
Stard10
|
StAR-related lipid transfer domain containing 10 |
| chr20_-_3751994 | 0.04 |
ENSRNOT00000072288
|
Pou5f1
|
POU class 5 homeobox 1 |
| chr7_-_144837583 | 0.04 |
ENSRNOT00000055289
|
Cbx5
|
chromobox 5 |
| chr3_-_23297774 | 0.04 |
ENSRNOT00000019162
|
Rpl35
|
ribosomal protein L35 |
| chr2_+_158097843 | 0.04 |
ENSRNOT00000016541
|
Ptx3
|
pentraxin 3 |
| chr13_+_51240911 | 0.04 |
ENSRNOT00000005551
|
Adipor1
|
adiponectin receptor 1 |
| chr2_+_147006830 | 0.04 |
ENSRNOT00000080780
|
AABR07010672.1
|
|
| chr9_+_97355924 | 0.04 |
ENSRNOT00000026558
|
Ackr3
|
atypical chemokine receptor 3 |
| chr18_-_29290447 | 0.04 |
ENSRNOT00000025353
|
Pfdn1
|
prefoldin subunit 1 |
| chr14_-_18862407 | 0.04 |
ENSRNOT00000003823
|
Cxcl6
|
C-X-C motif chemokine ligand 6 |
| chr1_+_282568287 | 0.04 |
ENSRNOT00000015997
|
Ces2i
|
carboxylesterase 2I |
| chr9_-_66627007 | 0.04 |
ENSRNOT00000049571
|
AABR07067829.1
|
|
| chr20_-_3299420 | 0.04 |
ENSRNOT00000090999
|
Gnl1
|
G protein nucleolar 1 |
| chr10_-_72556564 | 0.04 |
ENSRNOT00000048373
|
AABR07030162.1
|
|
| chr10_-_89088993 | 0.04 |
ENSRNOT00000027458
|
Ccr10
|
C-C motif chemokine receptor 10 |
| chr10_-_70788309 | 0.04 |
ENSRNOT00000029184
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr9_+_20213588 | 0.04 |
ENSRNOT00000089341
|
LOC100911515
|
triosephosphate isomerase-like |
| chr9_-_64096265 | 0.04 |
ENSRNOT00000013502
|
Tyw5
|
tRNA-yW synthesizing protein 5 |
| chr14_-_86739335 | 0.04 |
ENSRNOT00000078282
|
Purb
|
purine rich element binding protein B |
| chr9_-_3965939 | 0.04 |
ENSRNOT00000088289
|
AABR07066144.1
|
|
| chr2_+_86951776 | 0.04 |
ENSRNOT00000087275
|
AABR07009105.1
|
|
Network of associatons between targets according to the STRING database.
First level regulatory network of Rela_Rel_Nfkb1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.2 | 0.5 | GO:0070949 | regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) |
| 0.1 | 0.3 | GO:1903093 | regulation of protein K48-linked deubiquitination(GO:1903093) negative regulation of protein K48-linked deubiquitination(GO:1903094) negative regulation of ubiquitin-specific protease activity(GO:2000157) |
| 0.1 | 0.1 | GO:0033864 | positive regulation of NAD(P)H oxidase activity(GO:0033864) |
| 0.1 | 0.2 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0045210 | FasL biosynthetic process(GO:0045210) |
| 0.0 | 0.1 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0042376 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.0 | 0.1 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.2 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.2 | GO:0003105 | negative regulation of glomerular filtration(GO:0003105) |
| 0.0 | 0.1 | GO:0072144 | glomerular mesangial cell development(GO:0072144) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0042222 | interleukin-1 biosynthetic process(GO:0042222) |
| 0.0 | 0.0 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.1 | GO:0002458 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) negative regulation of interferon-gamma biosynthetic process(GO:0045077) |
| 0.0 | 0.1 | GO:0042509 | CD8-positive, alpha-beta T cell extravasation(GO:0035697) regulation of tyrosine phosphorylation of STAT protein(GO:0042509) positive regulation of tyrosine phosphorylation of STAT protein(GO:0042531) |
| 0.0 | 0.1 | GO:1990375 | baculum development(GO:1990375) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.4 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.3 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:0002901 | mature B cell apoptotic process(GO:0002901) regulation of mature B cell apoptotic process(GO:0002905) negative regulation of mature B cell apoptotic process(GO:0002906) |
| 0.0 | 0.0 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.0 | 0.0 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.0 | 0.1 | GO:0038158 | granulocyte colony-stimulating factor signaling pathway(GO:0038158) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.2 | GO:2000042 | negative regulation of double-strand break repair via homologous recombination(GO:2000042) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0036135 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.2 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.0 | 0.0 | GO:0002397 | MHC class I protein complex assembly(GO:0002397) |
| 0.0 | 0.1 | GO:1901423 | response to benzene(GO:1901423) |
| 0.0 | 0.0 | GO:0100012 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.0 | GO:0052203 | modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) regulation of exo-alpha-sialidase activity(GO:1903015) |
| 0.0 | 0.0 | GO:0030221 | basophil differentiation(GO:0030221) |
| 0.0 | 0.1 | GO:1902177 | positive regulation of oxidative stress-induced intrinsic apoptotic signaling pathway(GO:1902177) |
| 0.0 | 0.0 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:2000562 | negative regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000562) |
| 0.0 | 0.0 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.2 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.0 | GO:1903413 | cellular response to bile acid(GO:1903413) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) |
| 0.0 | 0.6 | GO:0070098 | chemokine-mediated signaling pathway(GO:0070098) |
| 0.0 | 0.0 | GO:0033123 | positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.0 | GO:0032902 | nerve growth factor production(GO:0032902) positive regulation of eating behavior(GO:1904000) |
| 0.0 | 0.0 | GO:0046968 | cytosol to ER transport(GO:0046967) peptide antigen transport(GO:0046968) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.1 | GO:1904706 | negative regulation of vascular smooth muscle cell proliferation(GO:1904706) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.2 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0042567 | insulin-like growth factor ternary complex(GO:0042567) |
| 0.0 | 0.1 | GO:0045098 | type III intermediate filament(GO:0045098) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036478 | tyrosine 3-monooxygenase activator activity(GO:0036470) L-dopa decarboxylase activator activity(GO:0036478) |
| 0.1 | 0.3 | GO:0043120 | tumor necrosis factor binding(GO:0043120) |
| 0.1 | 0.7 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0042903 | tubulin deacetylase activity(GO:0042903) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 1.0 | GO:0008139 | nuclear localization sequence binding(GO:0008139) |
| 0.0 | 0.1 | GO:0031752 | D5 dopamine receptor binding(GO:0031752) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.2 | GO:0016494 | C-X-C chemokine receptor activity(GO:0016494) |
| 0.0 | 0.2 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0004920 | interleukin-10 receptor activity(GO:0004920) |
| 0.0 | 0.0 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0031798 | type 1 metabotropic glutamate receptor binding(GO:0031798) |
| 0.0 | 0.0 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.0 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0070996 | type 1 melanocortin receptor binding(GO:0070996) |
| 0.0 | 0.1 | GO:0042289 | MHC class II protein binding(GO:0042289) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0035614 | snRNA stem-loop binding(GO:0035614) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.0 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.0 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0046979 | TAP binding(GO:0046977) TAP1 binding(GO:0046978) TAP2 binding(GO:0046979) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.0 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 0.3 | PID IL27 PATHWAY | IL27-mediated signaling events |
| 0.0 | 0.1 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.4 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.3 | PID IL23 PATHWAY | IL23-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME NFKB IS ACTIVATED AND SIGNALS SURVIVAL | Genes involved in NF-kB is activated and signals survival |
| 0.0 | 1.2 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.0 | REACTOME ACTIVATED TAK1 MEDIATES P38 MAPK ACTIVATION | Genes involved in activated TAK1 mediates p38 MAPK activation |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |