Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
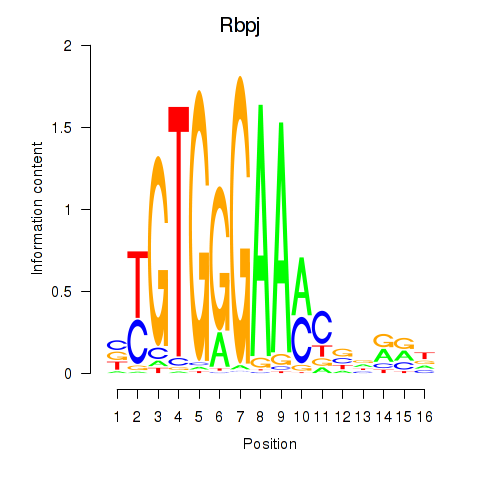
Results for Rbpj
Z-value: 0.33
Transcription factors associated with Rbpj
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rbpj
|
ENSRNOG00000007797 | recombination signal binding protein for immunoglobulin kappa J region |
|
Rbpj
|
ENSRNOG00000046327 | recombination signal binding protein for immunoglobulin kappa J region |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rbpj | rn6_v1_chr14_-_59735450_59735450 | 0.99 | 1.9e-03 | Click! |
Activity profile of Rbpj motif
Sorted Z-values of Rbpj motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_31890778 | 0.22 |
ENSRNOT00000039166
|
Tppp
|
tubulin polymerization promoting protein |
| chr19_+_755460 | 0.20 |
ENSRNOT00000076560
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr17_-_55709740 | 0.17 |
ENSRNOT00000033359
|
RGD1562037
|
similar to OTTHUMP00000046255 |
| chr18_+_27424328 | 0.15 |
ENSRNOT00000033784
|
Kif20a
|
kinesin family member 20A |
| chr5_+_172364421 | 0.13 |
ENSRNOT00000018769
|
Hes5
|
hes family bHLH transcription factor 5 |
| chr1_-_219412816 | 0.13 |
ENSRNOT00000083204
ENSRNOT00000029580 |
Rps6kb2
|
ribosomal protein S6 kinase B2 |
| chr7_+_11033317 | 0.12 |
ENSRNOT00000007498
|
Gna11
|
guanine nucleotide binding protein, alpha 11 |
| chrX_-_73778595 | 0.10 |
ENSRNOT00000076081
ENSRNOT00000075926 ENSRNOT00000003782 |
Rlim
|
ring finger protein, LIM domain interacting |
| chr1_+_84685931 | 0.10 |
ENSRNOT00000063839
|
Zfp780b
|
zinc finger protein 780B |
| chr3_-_141411170 | 0.10 |
ENSRNOT00000017364
|
Nkx2-2
|
NK2 homeobox 2 |
| chr9_+_94425252 | 0.09 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr18_+_56379890 | 0.09 |
ENSRNOT00000078764
|
Pdgfrb
|
platelet derived growth factor receptor beta |
| chr5_+_146383942 | 0.09 |
ENSRNOT00000078588
|
Csmd2
|
CUB and Sushi multiple domains 2 |
| chr20_+_5080070 | 0.09 |
ENSRNOT00000086950
ENSRNOT00000077082 |
Abhd16a
|
abhydrolase domain containing 16A |
| chr1_+_198214797 | 0.08 |
ENSRNOT00000068543
|
Tbx6
|
T-box 6 |
| chr14_+_34727915 | 0.08 |
ENSRNOT00000085089
|
Kdr
|
kinase insert domain receptor |
| chr3_-_112174269 | 0.08 |
ENSRNOT00000067836
|
Tmem87a
|
transmembrane protein 87A |
| chr17_+_76532611 | 0.08 |
ENSRNOT00000024099
|
Camk1d
|
calcium/calmodulin-dependent protein kinase ID |
| chr1_+_87697005 | 0.08 |
ENSRNOT00000028158
|
Zfp84
|
zinc finger protein 84 |
| chr3_-_9936352 | 0.08 |
ENSRNOT00000011224
ENSRNOT00000042798 |
Fnbp1
|
formin binding protein 1 |
| chr3_-_117990289 | 0.08 |
ENSRNOT00000011084
|
Shc4
|
SHC adaptor protein 4 |
| chr7_-_72328128 | 0.07 |
ENSRNOT00000008227
|
Tspyl5
|
TSPY-like 5 |
| chr11_+_81972219 | 0.07 |
ENSRNOT00000002452
|
Dgkg
|
diacylglycerol kinase, gamma |
| chr10_-_85636902 | 0.07 |
ENSRNOT00000017241
|
Pcgf2
|
polycomb group ring finger 2 |
| chr10_-_17075139 | 0.06 |
ENSRNOT00000039398
|
Neurl1b
|
neuralized E3 ubiquitin protein ligase 1B |
| chr8_+_114876146 | 0.06 |
ENSRNOT00000075966
|
Wdr82
|
WD repeat domain 82 |
| chr6_+_111049559 | 0.06 |
ENSRNOT00000015571
|
Tmem63c
|
transmembrane protein 63c |
| chr1_+_87790104 | 0.06 |
ENSRNOT00000074580
|
LOC103690114
|
zinc finger protein 383-like |
| chr18_-_27424090 | 0.06 |
ENSRNOT00000087968
|
Brd8
|
bromodomain containing 8 |
| chr16_+_78539489 | 0.06 |
ENSRNOT00000057897
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr17_-_22678253 | 0.06 |
ENSRNOT00000083122
|
LOC100362172
|
LRRGT00112-like |
| chr8_+_69971778 | 0.06 |
ENSRNOT00000058007
ENSRNOT00000037941 ENSRNOT00000050649 |
Megf11
|
multiple EGF-like-domains 11 |
| chr5_+_139468546 | 0.06 |
ENSRNOT00000013193
|
Slfnl1
|
schlafen-like 1 |
| chr13_-_73056765 | 0.06 |
ENSRNOT00000000049
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr8_-_61079526 | 0.05 |
ENSRNOT00000068658
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr11_-_87628502 | 0.05 |
ENSRNOT00000002568
|
Med15
|
mediator complex subunit 15 |
| chr8_+_119160731 | 0.05 |
ENSRNOT00000046745
|
Als2cl
|
ALS2 C-terminal like |
| chr2_+_184600721 | 0.05 |
ENSRNOT00000075823
|
Gatb
|
glutamyl-tRNA amidotransferase subunit B |
| chr11_-_70961358 | 0.05 |
ENSRNOT00000002427
|
Lmln
|
leishmanolysin like peptidase |
| chr5_-_144221263 | 0.05 |
ENSRNOT00000013570
|
Thrap3
|
thyroid hormone receptor associated protein 3 |
| chr10_-_15346236 | 0.05 |
ENSRNOT00000088104
ENSRNOT00000027430 |
Capn15
|
calpain 15 |
| chr2_-_178334900 | 0.05 |
ENSRNOT00000090152
|
Fnip2
|
folliculin interacting protein 2 |
| chr18_-_31071371 | 0.05 |
ENSRNOT00000060432
|
Diaph1
|
diaphanous-related formin 1 |
| chr13_+_110257571 | 0.05 |
ENSRNOT00000005715
|
Ints7
|
integrator complex subunit 7 |
| chr1_-_100671074 | 0.05 |
ENSRNOT00000027132
|
Myh14
|
myosin heavy chain 14 |
| chr10_+_55707164 | 0.04 |
ENSRNOT00000009757
|
Hes7
|
hes family bHLH transcription factor 7 |
| chr4_+_147037179 | 0.04 |
ENSRNOT00000011292
|
Syn2
|
synapsin II |
| chr1_-_88558696 | 0.04 |
ENSRNOT00000038642
|
AABR07002896.1
|
|
| chr6_+_107169528 | 0.04 |
ENSRNOT00000012495
|
Psen1
|
presenilin 1 |
| chr12_+_25450286 | 0.04 |
ENSRNOT00000092956
|
Gtf2i
|
general transcription factor II I |
| chr8_+_61659445 | 0.04 |
ENSRNOT00000023831
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr1_+_252894663 | 0.04 |
ENSRNOT00000054757
|
Ifit2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr12_-_38782010 | 0.04 |
ENSRNOT00000001813
|
Wdr66
|
WD repeat domain 66 |
| chr4_-_64831233 | 0.04 |
ENSRNOT00000079285
|
Dgki
|
diacylglycerol kinase, iota |
| chr5_+_135860675 | 0.04 |
ENSRNOT00000024737
|
Hectd3
|
HECT domain E3 ubiquitin protein ligase 3 |
| chr9_+_119517101 | 0.04 |
ENSRNOT00000020476
|
Lpin2
|
lipin 2 |
| chr12_+_19328957 | 0.04 |
ENSRNOT00000033288
|
Cnpy4
|
canopy FGF signaling regulator 4 |
| chr1_-_88920291 | 0.03 |
ENSRNOT00000028306
|
Kirrel2
|
kin of IRRE like 2 (Drosophila) |
| chr7_-_123088819 | 0.03 |
ENSRNOT00000056077
|
AC096601.1
|
|
| chr1_-_46978261 | 0.03 |
ENSRNOT00000060527
|
Serac1
|
serine active site containing 1 |
| chr3_+_8453609 | 0.03 |
ENSRNOT00000041921
|
Odf2
|
outer dense fiber of sperm tails 2 |
| chr2_+_114386019 | 0.03 |
ENSRNOT00000082148
|
AABR07009834.1
|
|
| chr1_+_91663736 | 0.03 |
ENSRNOT00000089587
ENSRNOT00000031236 |
Cep89
|
centrosomal protein 89 |
| chr14_+_4125380 | 0.03 |
ENSRNOT00000002882
|
Zfp644
|
zinc finger protein 644 |
| chr12_+_19314251 | 0.03 |
ENSRNOT00000001827
|
Ap4m1
|
adaptor-related protein complex 4, mu 1 subunit |
| chr1_+_185427736 | 0.03 |
ENSRNOT00000064706
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr17_-_45154355 | 0.03 |
ENSRNOT00000084976
|
Zkscan4
|
zinc finger with KRAB and SCAN domains 4 |
| chr3_+_150052483 | 0.03 |
ENSRNOT00000022265
|
RGD1561517
|
similar to chromosome 20 open reading frame 144 |
| chr14_+_34727623 | 0.03 |
ENSRNOT00000071405
ENSRNOT00000090183 |
Kdr
|
kinase insert domain receptor |
| chr5_+_82587420 | 0.03 |
ENSRNOT00000014020
|
Tlr4
|
toll-like receptor 4 |
| chr1_+_192115966 | 0.03 |
ENSRNOT00000025629
|
Plk1
|
polo-like kinase 1 |
| chr7_-_123088279 | 0.03 |
ENSRNOT00000071998
|
Tob2
|
transducer of ERBB2, 2 |
| chr1_-_226152524 | 0.03 |
ENSRNOT00000027756
|
Fads2
|
fatty acid desaturase 2 |
| chr9_+_81880177 | 0.03 |
ENSRNOT00000022839
|
Stk36
|
serine/threonine kinase 36 |
| chr18_+_30808404 | 0.03 |
ENSRNOT00000060475
|
Pcdhga1
|
protocadherin gamma subfamily A, 1 |
| chr12_-_29919320 | 0.03 |
ENSRNOT00000038092
|
Tyw1
|
tRNA-yW synthesizing protein 1 homolog |
| chr4_+_49017311 | 0.03 |
ENSRNOT00000007476
|
Ing3
|
inhibitor of growth family, member 3 |
| chr4_+_123118468 | 0.03 |
ENSRNOT00000010895
|
Tmem43
|
transmembrane protein 43 |
| chr6_+_110968061 | 0.02 |
ENSRNOT00000014980
|
Cipc
|
CLOCK-interacting pacemaker |
| chrX_+_68891227 | 0.02 |
ENSRNOT00000009635
|
Efnb1
|
ephrin B1 |
| chrX_+_14994016 | 0.02 |
ENSRNOT00000006365
|
Ftsj1
|
FtsJ RNA methyltransferase homolog 1 (E. coli) |
| chr1_+_84584596 | 0.02 |
ENSRNOT00000032933
|
LOC100910046
|
zinc finger protein 60-like |
| chr13_+_67611708 | 0.02 |
ENSRNOT00000063833
|
Tpr
|
translocated promoter region, nuclear basket protein |
| chr15_-_60766579 | 0.02 |
ENSRNOT00000079978
|
Akap11
|
A-kinase anchoring protein 11 |
| chr9_-_79545024 | 0.02 |
ENSRNOT00000021152
|
Mreg
|
melanoregulin |
| chr1_-_88428685 | 0.02 |
ENSRNOT00000074582
|
Zfp566
|
zinc finger protein 566 |
| chr5_-_129352886 | 0.02 |
ENSRNOT00000012088
|
Cdkn2c
|
cyclin-dependent kinase inhibitor 2C |
| chr2_-_236480502 | 0.02 |
ENSRNOT00000015020
|
Sgms2
|
sphingomyelin synthase 2 |
| chr7_+_116745061 | 0.02 |
ENSRNOT00000076174
|
Rhpn1
|
rhophilin, Rho GTPase binding protein 1 |
| chr1_-_88558387 | 0.02 |
ENSRNOT00000074174
|
AABR07002896.1
|
|
| chr1_-_261179790 | 0.02 |
ENSRNOT00000074420
ENSRNOT00000072073 |
Exosc1
|
exosome component 1 |
| chr19_+_61334168 | 0.02 |
ENSRNOT00000080465
|
Nrp1
|
neuropilin 1 |
| chr17_-_10061750 | 0.02 |
ENSRNOT00000082042
ENSRNOT00000022646 |
Zfp346
|
zinc finger protein 346 |
| chr2_-_233743866 | 0.02 |
ENSRNOT00000087062
|
Enpep
|
glutamyl aminopeptidase |
| chr7_-_135630654 | 0.02 |
ENSRNOT00000047388
ENSRNOT00000088223 ENSRNOT00000074793 |
Adamts20
|
ADAM metallopeptidase with thrombospondin type 1 motif, 20 |
| chr1_+_261180007 | 0.02 |
ENSRNOT00000072181
|
Zdhhc16
|
zinc finger, DHHC-type containing 16 |
| chr6_+_34094306 | 0.02 |
ENSRNOT00000051970
|
Wdr35
|
WD repeat domain 35 |
| chr10_+_103874383 | 0.02 |
ENSRNOT00000038935
|
Otop2
|
otopetrin 2 |
| chr2_+_166403265 | 0.02 |
ENSRNOT00000012417
|
Nmd3
|
NMD3 ribosome export adaptor |
| chr1_+_162342051 | 0.01 |
ENSRNOT00000016478
|
Alg8
|
ALG8, alpha-1,3-glucosyltransferase |
| chr1_+_29191192 | 0.01 |
ENSRNOT00000018718
|
Hey2
|
hes-related family bHLH transcription factor with YRPW motif 2 |
| chr12_-_19328637 | 0.01 |
ENSRNOT00000001829
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr1_-_87777359 | 0.01 |
ENSRNOT00000074090
|
AABR07002870.1
|
|
| chrX_+_110894845 | 0.01 |
ENSRNOT00000080072
|
Tbc1d8b
|
TBC1 domain family member 8B |
| chr12_-_48218955 | 0.01 |
ENSRNOT00000067975
ENSRNOT00000080557 ENSRNOT00000000821 |
Acacb
|
acetyl-CoA carboxylase beta |
| chr6_-_30187337 | 0.01 |
ENSRNOT00000074150
|
Itsn2
|
intersectin 2 |
| chr20_+_3556560 | 0.01 |
ENSRNOT00000085635
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr7_+_2635743 | 0.01 |
ENSRNOT00000004223
|
Mip
|
major intrinsic protein of lens fiber |
| chr3_+_7279340 | 0.01 |
ENSRNOT00000017029
|
Ak8
|
adenylate kinase 8 |
| chr7_-_137856485 | 0.01 |
ENSRNOT00000007003
|
LOC688906
|
similar to splicing factor, arginine/serine-rich 2, interacting protein |
| chr10_+_39666991 | 0.01 |
ENSRNOT00000030760
|
Acsl6
|
acyl-CoA synthetase long-chain family member 6 |
| chr3_+_37599728 | 0.01 |
ENSRNOT00000090575
|
Rif1
|
replication timing regulatory factor 1 |
| chr19_+_57820260 | 0.01 |
ENSRNOT00000057945
|
Disc1
|
disrupted in schizophrenia 1 |
| chr1_+_23977688 | 0.01 |
ENSRNOT00000014805
|
Tbpl1
|
TATA-box binding protein like 1 |
| chr1_+_199412834 | 0.01 |
ENSRNOT00000031891
|
Fus
|
fused in sarcoma RNA binding protein |
| chr3_-_3594475 | 0.01 |
ENSRNOT00000064861
|
RGD1564379
|
RGD1564379 |
| chr3_-_104502471 | 0.01 |
ENSRNOT00000040306
|
Ryr3
|
ryanodine receptor 3 |
| chr1_+_79931156 | 0.01 |
ENSRNOT00000019814
ENSRNOT00000084985 |
Sympk
|
symplekin |
| chrX_-_112473822 | 0.01 |
ENSRNOT00000079180
|
Col4a6
|
collagen type IV alpha 6 chain |
| chr1_+_157573324 | 0.01 |
ENSRNOT00000092066
|
Rab30
|
RAB30, member RAS oncogene family |
| chr3_+_161212156 | 0.01 |
ENSRNOT00000020280
ENSRNOT00000083553 |
Dnttip1
|
deoxynucleotidyltransferase, terminal, interacting protein 1 |
| chr12_-_23727535 | 0.01 |
ENSRNOT00000085911
ENSRNOT00000001950 |
Dtx2
|
deltex E3 ubiquitin ligase 2 |
| chr1_-_101008266 | 0.01 |
ENSRNOT00000081430
|
Scaf1
|
SR-related CTD-associated factor 1 |
| chrX_-_15651332 | 0.01 |
ENSRNOT00000039000
|
Gpkow
|
G patch domain and KOW motifs |
| chr18_-_37245809 | 0.01 |
ENSRNOT00000079585
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr1_-_189181901 | 0.01 |
ENSRNOT00000092022
|
Gp2
|
glycoprotein 2 |
| chr16_-_74864816 | 0.01 |
ENSRNOT00000017164
|
Alg11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr4_+_67059939 | 0.01 |
ENSRNOT00000052393
|
Chmp4bl1
|
chromatin modifying protein 4B-like 1 |
| chr3_+_3834268 | 0.01 |
ENSRNOT00000031622
|
Pmpca
|
peptidase, mitochondrial processing alpha subunit |
| chr6_+_64252020 | 0.00 |
ENSRNOT00000047296
ENSRNOT00000082105 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr7_-_33584564 | 0.00 |
ENSRNOT00000005463
|
Nedd1
|
neural precursor cell expressed, developmentally down-regulated 1 |
| chr12_+_22142520 | 0.00 |
ENSRNOT00000038397
|
Fbxo24
|
F-box protein 24 |
| chr7_+_123043503 | 0.00 |
ENSRNOT00000026258
ENSRNOT00000086355 |
Tef
|
TEF, PAR bZIP transcription factor |
| chr3_+_45683993 | 0.00 |
ENSRNOT00000038983
|
Tanc1
|
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 |
| chr8_+_116657371 | 0.00 |
ENSRNOT00000024966
|
Mon1a
|
MON1 homolog A, secretory trafficking associated |
| chr8_-_57830504 | 0.00 |
ENSRNOT00000017683
|
Ddx10
|
DEAD-box helicase 10 |
| chr2_-_196456481 | 0.00 |
ENSRNOT00000028677
|
Prune
|
prune exopolyphosphatase |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rbpj
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) |
| 0.0 | 0.1 | GO:0060816 | random inactivation of X chromosome(GO:0060816) regulation of dosage compensation by inactivation of X chromosome(GO:1900095) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0072277 | cell migration involved in vasculogenesis(GO:0035441) VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.1 | GO:0048597 | positive regulation of endothelial cell chemotaxis by VEGF-activated vascular endothelial growth factor receptor signaling pathway(GO:0038033) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.1 | GO:0060158 | phospholipase C-activating dopamine receptor signaling pathway(GO:0060158) |
| 0.0 | 0.1 | GO:0060267 | positive regulation of respiratory burst(GO:0060267) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.0 | GO:0097513 | myosin II filament(GO:0097513) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031826 | type 2A serotonin receptor binding(GO:0031826) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.1 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.0 | 0.1 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.0 | 0.1 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.1 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |