Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
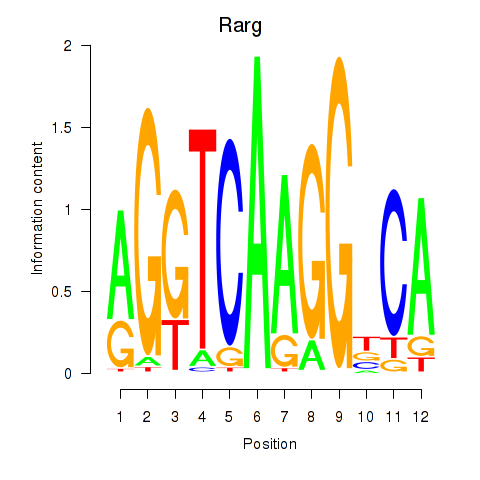
Results for Rarg
Z-value: 0.54
Transcription factors associated with Rarg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rarg
|
ENSRNOG00000012499 | retinoic acid receptor, gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rarg | rn6_v1_chr7_-_143852119_143852119 | -0.80 | 1.0e-01 | Click! |
Activity profile of Rarg motif
Sorted Z-values of Rarg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_123106694 | 1.31 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr3_-_123119460 | 0.49 |
ENSRNOT00000028833
|
Avp
|
arginine vasopressin |
| chr12_-_40227297 | 0.30 |
ENSRNOT00000030600
|
Fam109a
|
family with sequence similarity 109, member A |
| chr11_+_30904733 | 0.30 |
ENSRNOT00000036027
|
Mrap
|
melanocortin 2 receptor accessory protein |
| chr4_+_148782479 | 0.28 |
ENSRNOT00000018133
|
LOC500300
|
similar to hypothetical protein MGC6835 |
| chr10_-_15603649 | 0.23 |
ENSRNOT00000051483
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr16_+_20672374 | 0.22 |
ENSRNOT00000063780
|
RGD1566239
|
similar to RIKEN cDNA 2810428I15 |
| chr10_-_90307658 | 0.22 |
ENSRNOT00000092102
|
Slc4a1
|
solute carrier family 4 member 1 |
| chr4_-_119889949 | 0.20 |
ENSRNOT00000033687
|
H1fx
|
H1 histone family, member X |
| chr4_+_169161585 | 0.20 |
ENSRNOT00000079785
|
Emp1
|
epithelial membrane protein 1 |
| chr1_-_80744831 | 0.19 |
ENSRNOT00000025913
|
Bcl3
|
B-cell CLL/lymphoma 3 |
| chr3_-_120306551 | 0.18 |
ENSRNOT00000021082
|
Mall
|
mal, T-cell differentiation protein-like |
| chr19_+_55917736 | 0.17 |
ENSRNOT00000020635
|
Rpl13
|
ribosomal protein L13 |
| chr1_-_81450094 | 0.17 |
ENSRNOT00000035356
|
Zfp575
|
zinc finger protein 575 |
| chr1_+_101412736 | 0.16 |
ENSRNOT00000067468
|
Lhb
|
luteinizing hormone beta polypeptide |
| chr1_+_106998623 | 0.16 |
ENSRNOT00000022383
|
Slc17a6
|
solute carrier family 17 member 6 |
| chr1_-_226049929 | 0.16 |
ENSRNOT00000007320
|
Best1
|
bestrophin 1 |
| chr4_-_165629996 | 0.16 |
ENSRNOT00000068185
ENSRNOT00000007427 |
Ybx3
|
Y box binding protein 3 |
| chr15_-_4035064 | 0.15 |
ENSRNOT00000012300
|
Fut11
|
fucosyltransferase 11 |
| chr20_-_13657943 | 0.15 |
ENSRNOT00000032290
|
Vpreb3
|
pre-B lymphocyte 3 |
| chr20_-_5618254 | 0.15 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr8_+_118662335 | 0.14 |
ENSRNOT00000029755
|
Ngp
|
neutrophilic granule protein |
| chrX_-_118998300 | 0.14 |
ENSRNOT00000025575
|
AABR07041089.1
|
|
| chr7_-_11513415 | 0.14 |
ENSRNOT00000082486
|
Thop1
|
thimet oligopeptidase 1 |
| chr1_+_90919076 | 0.14 |
ENSRNOT00000081362
|
LOC108348197
|
calpain small subunit 1-like |
| chr10_-_63176463 | 0.14 |
ENSRNOT00000004717
|
Slc6a4
|
solute carrier family 6 member 4 |
| chr4_-_181477281 | 0.14 |
ENSRNOT00000055463
|
Mansc4
|
MANSC domain containing 4 |
| chr4_+_153774486 | 0.13 |
ENSRNOT00000074096
|
Tuba8
|
tubulin, alpha 8 |
| chr12_+_24989298 | 0.13 |
ENSRNOT00000032780
|
Eln
|
elastin |
| chr13_+_52588917 | 0.13 |
ENSRNOT00000011999
|
Phlda3
|
pleckstrin homology-like domain, family A, member 3 |
| chr7_-_12646960 | 0.13 |
ENSRNOT00000014687
|
Prtn3
|
proteinase 3 |
| chr6_-_136379348 | 0.13 |
ENSRNOT00000016208
|
Xrcc3
|
X-ray repair cross complementing 3 |
| chr4_+_165732643 | 0.12 |
ENSRNOT00000034403
|
LOC690326
|
hypothetical protein LOC690326 |
| chr10_-_56429748 | 0.12 |
ENSRNOT00000020675
ENSRNOT00000092704 |
Spem1
|
spermatid maturation 1 |
| chr20_-_4489281 | 0.12 |
ENSRNOT00000031548
|
Cyp21a1
|
cytochrome P450, family 21, subfamily a, polypeptide 1 |
| chr1_+_78876205 | 0.12 |
ENSRNOT00000022610
|
Pnmal2
|
paraneoplastic Ma antigen family-like 2 |
| chr12_-_2007516 | 0.12 |
ENSRNOT00000037564
|
Pex11g
|
peroxisomal biogenesis factor 11 gamma |
| chr10_+_55707164 | 0.12 |
ENSRNOT00000009757
|
Hes7
|
hes family bHLH transcription factor 7 |
| chr7_-_11018160 | 0.12 |
ENSRNOT00000092061
|
Aes
|
amino-terminal enhancer of split |
| chr10_-_40805941 | 0.11 |
ENSRNOT00000017588
|
Atox1
|
antioxidant 1 copper chaperone |
| chr20_+_3226360 | 0.11 |
ENSRNOT00000070903
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr20_-_13706205 | 0.11 |
ENSRNOT00000038623
|
Derl3
|
derlin 3 |
| chr2_+_198417619 | 0.11 |
ENSRNOT00000085945
|
Hist2h3c2
|
histone cluster 2, H3c2 |
| chrX_+_107496072 | 0.11 |
ENSRNOT00000003283
|
Plp1
|
proteolipid protein 1 |
| chr20_+_44680449 | 0.11 |
ENSRNOT00000000728
|
Traf3ip2
|
Traf3 interacting protein 2 |
| chr4_-_123118186 | 0.11 |
ENSRNOT00000038096
|
LOC100361898
|
coiled-coil-helix-coiled-coil-helix domain containing 4 |
| chr6_+_146784915 | 0.11 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr6_+_136358057 | 0.11 |
ENSRNOT00000092741
|
Klc1
|
kinesin light chain 1 |
| chr10_-_88677055 | 0.10 |
ENSRNOT00000025590
|
Ghdc
|
GH3 domain containing |
| chr10_+_38889370 | 0.10 |
ENSRNOT00000091236
|
Sept8
|
septin 8 |
| chr7_-_11513581 | 0.10 |
ENSRNOT00000027045
|
Thop1
|
thimet oligopeptidase 1 |
| chr1_-_261446570 | 0.10 |
ENSRNOT00000020182
|
Sfrp5
|
secreted frizzled-related protein 5 |
| chr8_+_22047697 | 0.09 |
ENSRNOT00000067741
|
Icam4
|
intercellular adhesion molecule 4, Landsteiner-Wiener blood group |
| chr5_+_8916633 | 0.09 |
ENSRNOT00000066395
|
Ppp1r42
|
protein phosphatase 1, regulatory subunit 42 |
| chr2_-_53718143 | 0.09 |
ENSRNOT00000049363
|
AABR07008350.1
|
|
| chr9_-_17254949 | 0.09 |
ENSRNOT00000026499
|
Mrps18a
|
mitochondrial ribosomal protein S18A |
| chr20_+_31234493 | 0.09 |
ENSRNOT00000000675
|
Npffr1
|
neuropeptide FF receptor 1 |
| chr7_+_72924799 | 0.09 |
ENSRNOT00000008969
|
Laptm4b
|
lysosomal protein transmembrane 4 beta |
| chr4_+_168832910 | 0.09 |
ENSRNOT00000011134
|
Gprc5a
|
G protein-coupled receptor, class C, group 5, member A |
| chr7_-_118518193 | 0.09 |
ENSRNOT00000075636
|
LOC102555083
|
zinc finger protein 250-like |
| chr10_+_10774639 | 0.09 |
ENSRNOT00000061208
|
Sept12
|
septin 12 |
| chr4_-_100783750 | 0.09 |
ENSRNOT00000078956
|
Kcmf1
|
potassium channel modulatory factor 1 |
| chr9_+_9721105 | 0.09 |
ENSRNOT00000073042
ENSRNOT00000075494 |
C3
|
complement C3 |
| chr7_-_121489078 | 0.09 |
ENSRNOT00000092529
|
Rps19bp1
|
ribosomal protein S19 binding protein 1 |
| chr2_+_195603599 | 0.09 |
ENSRNOT00000028263
|
C2cd4d
|
C2 calcium-dependent domain containing 4D |
| chr8_-_66863476 | 0.09 |
ENSRNOT00000018820
|
Rplp1
|
ribosomal protein lateral stalk subunit P1 |
| chr7_+_13062196 | 0.08 |
ENSRNOT00000000193
|
Plpp2
|
phospholipid phosphatase 2 |
| chr1_-_67390141 | 0.08 |
ENSRNOT00000025808
|
Sbk1
|
SH3 domain binding kinase 1 |
| chr13_+_85918252 | 0.08 |
ENSRNOT00000006163
|
Lmx1a
|
LIM homeobox transcription factor 1 alpha |
| chr10_+_66099531 | 0.08 |
ENSRNOT00000056192
|
Lyrm9
|
LYR motif containing 9 |
| chr20_+_4329811 | 0.08 |
ENSRNOT00000000513
|
Notch4
|
notch 4 |
| chr4_+_99239115 | 0.08 |
ENSRNOT00000009515
|
Cd8a
|
CD8a molecule |
| chr10_-_91448477 | 0.08 |
ENSRNOT00000038836
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr7_-_115979298 | 0.08 |
ENSRNOT00000075883
|
RGD1308195
|
similar to secreted Ly6/uPAR related protein 2 |
| chr1_+_78015722 | 0.08 |
ENSRNOT00000002043
|
Kptn
|
kaptin (actin binding protein) |
| chr8_-_56393233 | 0.08 |
ENSRNOT00000016263
|
Fdx1
|
ferredoxin 1 |
| chr8_-_23014499 | 0.08 |
ENSRNOT00000017820
|
Ccdc151
|
coiled-coil domain containing 151 |
| chr1_-_89539210 | 0.08 |
ENSRNOT00000077462
ENSRNOT00000028644 |
Hpn
|
hepsin |
| chr1_-_79899648 | 0.08 |
ENSRNOT00000057960
|
AC110846.1
|
|
| chr1_-_166943592 | 0.08 |
ENSRNOT00000026962
|
Folr1
|
folate receptor 1 |
| chr5_+_160306727 | 0.08 |
ENSRNOT00000016648
|
Agmat
|
agmatinase |
| chr12_-_13668515 | 0.08 |
ENSRNOT00000086847
|
Fscn1
|
fascin actin-bundling protein 1 |
| chr20_+_5008508 | 0.08 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr16_-_20148867 | 0.08 |
ENSRNOT00000040464
|
Fcho1
|
FCH domain only 1 |
| chr9_+_10956056 | 0.08 |
ENSRNOT00000073930
|
Plin5
|
perilipin 5 |
| chr16_+_21288876 | 0.08 |
ENSRNOT00000027990
|
Cilp2
|
cartilage intermediate layer protein 2 |
| chr3_+_79613171 | 0.08 |
ENSRNOT00000011467
|
Agbl2
|
ATP/GTP binding protein-like 2 |
| chr9_-_119332967 | 0.08 |
ENSRNOT00000021048
|
Myl12a
|
myosin light chain 12A |
| chr1_+_219250265 | 0.08 |
ENSRNOT00000024353
|
Doc2g
|
double C2-like domains, gamma |
| chr9_-_9985630 | 0.08 |
ENSRNOT00000071780
|
Crb3
|
crumbs 3, cell polarity complex component |
| chr9_+_73493027 | 0.08 |
ENSRNOT00000074427
ENSRNOT00000089478 |
Unc80
|
unc-80 homolog, NALCN activator |
| chrX_+_106487870 | 0.08 |
ENSRNOT00000075521
|
Arxes2
|
adipocyte-related X-chromosome expressed sequence 2 |
| chr15_+_44441856 | 0.08 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr11_-_34142753 | 0.08 |
ENSRNOT00000002297
|
Cldn14
|
claudin 14 |
| chr6_+_137997335 | 0.07 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr19_+_55094585 | 0.07 |
ENSRNOT00000068452
|
Zfpm1
|
zinc finger protein, multitype 1 |
| chr7_-_29152442 | 0.07 |
ENSRNOT00000079774
|
Mybpc1
|
myosin binding protein C, slow type |
| chr8_-_128711221 | 0.07 |
ENSRNOT00000055888
|
Xirp1
|
xin actin-binding repeat containing 1 |
| chr7_-_144960527 | 0.07 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr1_+_81396558 | 0.07 |
ENSRNOT00000081829
|
Irgq
|
immunity-related GTPase Q |
| chr9_-_121713091 | 0.07 |
ENSRNOT00000073432
|
Adcyap1
|
adenylate cyclase activating polypeptide 1 |
| chr7_+_120176530 | 0.07 |
ENSRNOT00000087548
|
Triobp
|
TRIO and F-actin binding protein |
| chr4_-_145454834 | 0.07 |
ENSRNOT00000013154
ENSRNOT00000056493 |
Prrt3
|
proline-rich transmembrane protein 3 |
| chr7_-_117978787 | 0.07 |
ENSRNOT00000074262
|
Zfp647
|
zinc finger protein 647 |
| chr10_-_90999506 | 0.07 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr11_+_64472072 | 0.07 |
ENSRNOT00000042756
|
RGD1563835
|
similar to ribosomal protein L27 |
| chr1_+_221801524 | 0.07 |
ENSRNOT00000031227
|
Nrxn2
|
neurexin 2 |
| chr17_-_13812615 | 0.07 |
ENSRNOT00000019473
ENSRNOT00000085581 |
S1pr3
|
sphingosine-1-phosphate receptor 3 |
| chr8_+_48569328 | 0.07 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr8_-_63092009 | 0.07 |
ENSRNOT00000034300
|
Loxl1
|
lysyl oxidase-like 1 |
| chr13_-_91228901 | 0.07 |
ENSRNOT00000071728
ENSRNOT00000073643 ENSRNOT00000071897 |
LOC100911825
|
low affinity immunoglobulin gamma Fc region receptor III-like |
| chr12_+_22153983 | 0.07 |
ENSRNOT00000080775
ENSRNOT00000001894 |
Pcolce
|
procollagen C-endopeptidase enhancer |
| chr7_+_11769400 | 0.07 |
ENSRNOT00000044417
|
Jsrp1
|
junctional sarcoplasmic reticulum protein 1 |
| chr5_-_140657745 | 0.07 |
ENSRNOT00000019080
|
Mfsd2a
|
major facilitator superfamily domain containing 2A |
| chr17_+_9282675 | 0.07 |
ENSRNOT00000051702
|
H2afy
|
H2A histone family, member Y |
| chr18_-_16542165 | 0.07 |
ENSRNOT00000079381
|
Slc39a6
|
solute carrier family 39 member 6 |
| chr10_-_105771972 | 0.07 |
ENSRNOT00000074286
ENSRNOT00000066137 |
Mxra7
|
matrix remodeling associated 7 |
| chr5_+_152290084 | 0.07 |
ENSRNOT00000089849
|
Aim1l
|
absent in melanoma 1-like |
| chr19_+_320906 | 0.06 |
ENSRNOT00000041664
|
AABR07042611.1
|
|
| chr19_+_14346197 | 0.06 |
ENSRNOT00000043480
|
Gm18025
|
predicted gene, 18025 |
| chr10_+_103206014 | 0.06 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chrX_-_123662350 | 0.06 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr7_+_120755516 | 0.06 |
ENSRNOT00000056169
|
Kdelr3
|
KDEL endoplasmic reticulum protein retention receptor 3 |
| chr1_-_89303968 | 0.06 |
ENSRNOT00000056714
|
Ffar3
|
free fatty acid receptor 3 |
| chr12_+_13323547 | 0.06 |
ENSRNOT00000074138
|
Zfp853
|
zinc finger protein 853 |
| chr20_+_7409401 | 0.06 |
ENSRNOT00000000586
|
Snrpc
|
small nuclear ribonucleoprotein polypeptide C |
| chr1_+_81763614 | 0.06 |
ENSRNOT00000027254
|
Cd79a
|
CD79a molecule |
| chr4_-_30338679 | 0.06 |
ENSRNOT00000012050
|
Pon3
|
paraoxonase 3 |
| chr10_+_89635675 | 0.06 |
ENSRNOT00000028179
|
RGD1561590
|
similar to SAP18 |
| chr8_+_96564877 | 0.06 |
ENSRNOT00000017879
|
Mthfs
|
5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) |
| chr2_-_183128932 | 0.06 |
ENSRNOT00000031179
|
Mnd1
|
meiotic nuclear divisions 1 |
| chr2_-_257376756 | 0.06 |
ENSRNOT00000065811
|
Gipc2
|
GIPC PDZ domain containing family, member 2 |
| chr13_+_75105615 | 0.06 |
ENSRNOT00000076653
|
Tp53i3
|
tumor protein p53 inducible protein 3 |
| chr15_+_34584320 | 0.06 |
ENSRNOT00000048255
|
AABR07017917.1
|
|
| chr2_+_23385183 | 0.06 |
ENSRNOT00000014860
|
Arsb
|
arylsulfatase B |
| chr3_+_168124673 | 0.06 |
ENSRNOT00000070809
|
Pfdn4
|
prefoldin subunit 4 |
| chr6_-_6842758 | 0.06 |
ENSRNOT00000006094
|
Kcng3
|
potassium voltage-gated channel modifier subfamily G member 3 |
| chr8_-_12902262 | 0.06 |
ENSRNOT00000033969
|
Endod1
|
endonuclease domain containing 1 |
| chrX_+_15273933 | 0.06 |
ENSRNOT00000075082
|
LOC108348091
|
erythroid transcription factor |
| chr1_-_102255459 | 0.06 |
ENSRNOT00000067392
|
Ush1c
|
USH1 protein network component harmonin |
| chr19_-_37528011 | 0.06 |
ENSRNOT00000059628
|
Agrp
|
agouti related neuropeptide |
| chr8_+_50604768 | 0.06 |
ENSRNOT00000059383
|
AC135409.1
|
|
| chr20_-_32139789 | 0.06 |
ENSRNOT00000078140
|
Srgn
|
serglycin |
| chr3_+_1452644 | 0.06 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr4_+_181434812 | 0.06 |
ENSRNOT00000002520
|
Mrps35
|
mitochondrial ribosomal protein S35 |
| chr11_-_88177448 | 0.06 |
ENSRNOT00000085475
|
Ypel1
|
yippee-like 1 |
| chr6_-_91490366 | 0.06 |
ENSRNOT00000059187
|
Dnaaf2
|
dynein, axonemal, assembly factor 2 |
| chr2_+_197682000 | 0.06 |
ENSRNOT00000066821
|
Hormad1
|
HORMA domain containing 1 |
| chr12_+_2046472 | 0.06 |
ENSRNOT00000001289
|
Zfp358
|
zinc finger protein 358 |
| chr7_+_121889157 | 0.06 |
ENSRNOT00000024835
|
Fam83f
|
family with sequence similarity 83, member F |
| chr10_-_66020682 | 0.06 |
ENSRNOT00000011019
|
Fam58b
|
family with sequence similarity 58, member B |
| chr10_-_57275708 | 0.06 |
ENSRNOT00000005370
|
Pfn1
|
profilin 1 |
| chr19_+_37795658 | 0.06 |
ENSRNOT00000025686
|
Tsnaxip1
|
translin-associated factor X interacting protein 1 |
| chr10_-_18131562 | 0.06 |
ENSRNOT00000006676
|
Tlx3
|
T-cell leukemia, homeobox 3 |
| chr14_-_17333588 | 0.05 |
ENSRNOT00000077636
|
Ppef2
|
protein phosphatase with EF-hand domain 2 |
| chr10_+_105498728 | 0.05 |
ENSRNOT00000032163
|
Sphk1
|
sphingosine kinase 1 |
| chr2_+_200572502 | 0.05 |
ENSRNOT00000074666
|
Zfp697
|
zinc finger protein 697 |
| chr1_-_88558387 | 0.05 |
ENSRNOT00000074174
|
AABR07002896.1
|
|
| chr5_-_150484207 | 0.05 |
ENSRNOT00000090736
|
Rab42
|
RAB42, member RAS oncogene family |
| chr4_-_28953067 | 0.05 |
ENSRNOT00000013989
|
Tfpi2
|
tissue factor pathway inhibitor 2 |
| chr18_+_28653592 | 0.05 |
ENSRNOT00000085120
ENSRNOT00000045795 |
Cxxc5
|
CXXC finger protein 5 |
| chr16_+_20668971 | 0.05 |
ENSRNOT00000027073
|
Uba52
|
ubiquitin A-52 residue ribosomal protein fusion product 1 |
| chr1_+_97632473 | 0.05 |
ENSRNOT00000023671
|
Vstm2b
|
V-set and transmembrane domain containing 2B |
| chr1_+_100473643 | 0.05 |
ENSRNOT00000026379
|
Josd2
|
Josephin domain containing 2 |
| chr1_-_53014466 | 0.05 |
ENSRNOT00000049831
|
Sft2d1
|
SFT2 domain containing 1 |
| chr10_+_83476107 | 0.05 |
ENSRNOT00000075733
|
Phb
|
prohibitin |
| chr16_+_71242470 | 0.05 |
ENSRNOT00000030489
|
Letm2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr3_+_2643610 | 0.05 |
ENSRNOT00000037169
|
Fut7
|
fucosyltransferase 7 |
| chr10_+_39066716 | 0.05 |
ENSRNOT00000010729
|
Il5
|
interleukin 5 |
| chr16_-_20652889 | 0.05 |
ENSRNOT00000077756
|
Fkbp8
|
FK506 binding protein 8 |
| chr13_+_78805347 | 0.05 |
ENSRNOT00000003748
|
Serpinc1
|
serpin family C member 1 |
| chr10_+_82292110 | 0.05 |
ENSRNOT00000004435
|
Chad
|
chondroadherin |
| chr7_+_121311024 | 0.05 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr10_+_71202456 | 0.05 |
ENSRNOT00000076893
|
Hnf1b
|
HNF1 homeobox B |
| chr4_+_98027554 | 0.05 |
ENSRNOT00000009503
|
Serbp1
|
Serpine1 mRNA binding protein 1 |
| chr8_+_103337472 | 0.05 |
ENSRNOT00000049729
|
Paqr9
|
progestin and adipoQ receptor family member 9 |
| chr11_+_65960277 | 0.05 |
ENSRNOT00000003674
|
Ndufb4
|
NADH:ubiquinone oxidoreductase subunit B4 |
| chr1_-_91042230 | 0.05 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chr1_-_222177421 | 0.05 |
ENSRNOT00000078393
|
Esrra
|
estrogen related receptor, alpha |
| chr12_-_30566032 | 0.05 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr4_-_155051429 | 0.05 |
ENSRNOT00000020094
|
Klrg1
|
killer cell lectin like receptor G1 |
| chr8_+_116307141 | 0.05 |
ENSRNOT00000037375
|
Rassf1
|
Ras association domain family member 1 |
| chr9_-_71900044 | 0.05 |
ENSRNOT00000020322
|
Idh1
|
isocitrate dehydrogenase (NADP(+)) 1, cytosolic |
| chr20_+_5057701 | 0.05 |
ENSRNOT00000001119
|
Ly6g6c
|
lymphocyte antigen 6 complex, locus G6C |
| chr13_-_52197205 | 0.05 |
ENSRNOT00000009712
|
Shisa4
|
shisa family member 4 |
| chr2_-_80293181 | 0.05 |
ENSRNOT00000016111
|
Otulin
|
OTU deubiquitinase with linear linkage specificity |
| chr2_+_210880777 | 0.05 |
ENSRNOT00000026237
|
Gnat2
|
G protein subunit alpha transducin 2 |
| chr7_+_98813040 | 0.05 |
ENSRNOT00000012616
|
Ndufb9
|
NADH:ubiquinone oxidoreductase subunit B9 |
| chr13_-_72890841 | 0.05 |
ENSRNOT00000093726
|
RGD1304622
|
similar to 6820428L09 protein |
| chr5_+_139963002 | 0.05 |
ENSRNOT00000048506
|
Col9a2
|
collagen type IX alpha 2 chain |
| chr2_+_38230757 | 0.05 |
ENSRNOT00000048598
|
RGD1564606
|
similar to 60S ribosomal protein L23a |
| chr1_-_89360733 | 0.05 |
ENSRNOT00000028544
|
Mag
|
myelin-associated glycoprotein |
| chr9_-_85626094 | 0.05 |
ENSRNOT00000020919
|
Serpine2
|
serpin family E member 2 |
| chr4_-_30556814 | 0.05 |
ENSRNOT00000012760
|
Pdk4
|
pyruvate dehydrogenase kinase 4 |
| chr20_-_5140304 | 0.05 |
ENSRNOT00000092646
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr9_-_16795887 | 0.05 |
ENSRNOT00000071609
|
Cd79al
|
Cd79a molecule-like |
| chr12_-_24556976 | 0.05 |
ENSRNOT00000041798
|
Bcl7b
|
BCL tumor suppressor 7B |
| chr10_-_29026002 | 0.05 |
ENSRNOT00000005070
|
Pttg1
|
pituitary tumor-transforming 1 |
| chr15_-_34444244 | 0.05 |
ENSRNOT00000027612
|
Cideb
|
cell death-inducing DFFA-like effector b |
| chr1_-_199624783 | 0.05 |
ENSRNOT00000026908
|
Cox6a2
|
cytochrome c oxidase subunit 6A2 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rarg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.8 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.1 | 0.2 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0002877 | regulation of acute inflammatory response to non-antigenic stimulus(GO:0002877) |
| 0.0 | 0.1 | GO:0071298 | cellular response to L-ascorbic acid(GO:0071298) |
| 0.0 | 0.2 | GO:0010037 | response to carbon dioxide(GO:0010037) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0045041 | protein import into mitochondrial intermembrane space(GO:0045041) |
| 0.0 | 0.1 | GO:2000259 | positive regulation of complement activation(GO:0045917) positive regulation of protein activation cascade(GO:2000259) |
| 0.0 | 0.1 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:1903849 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.0 | 0.1 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.0 | 0.2 | GO:0001766 | membrane raft polarization(GO:0001766) membrane raft distribution(GO:0031580) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0042097 | mitral valve formation(GO:0003192) interleukin-4 biosynthetic process(GO:0042097) regulation of interleukin-4 biosynthetic process(GO:0045402) transcriptional activation by promoter-enhancer looping(GO:0071733) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0048597 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:0030222 | eosinophil differentiation(GO:0030222) |
| 0.0 | 0.1 | GO:1904714 | regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.0 | 0.1 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.2 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.0 | 0.1 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:1902626 | assembly of large subunit precursor of preribosome(GO:1902626) |
| 0.0 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.0 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:1904401 | response to Thyroid stimulating hormone(GO:1904400) cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.0 | 0.1 | GO:0042663 | regulation of pronephros size(GO:0035565) regulation of endodermal cell fate specification(GO:0042663) hepatoblast differentiation(GO:0061017) |
| 0.0 | 0.1 | GO:1990379 | lipid transport across blood brain barrier(GO:1990379) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.0 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.0 | 0.0 | GO:0100012 | regulation of heart induction by regulation of canonical Wnt signaling pathway(GO:0090081) regulation of heart induction(GO:0090381) regulation of heart induction by canonical Wnt signaling pathway(GO:0100012) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.2 | GO:0036065 | fucosylation(GO:0036065) |
| 0.0 | 0.0 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.0 | GO:1901227 | negative regulation of transcription from RNA polymerase II promoter involved in heart development(GO:1901227) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.0 | GO:1990375 | baculum development(GO:1990375) |
| 0.0 | 0.1 | GO:0046487 | glyoxylate metabolic process(GO:0046487) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.0 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.0 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:1904970 | brush border assembly(GO:1904970) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.0 | GO:0071895 | odontoblast differentiation(GO:0071895) |
| 0.0 | 0.1 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.0 | GO:0002434 | immune complex clearance(GO:0002434) |
| 0.0 | 0.0 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:0042822 | pyridoxal phosphate metabolic process(GO:0042822) |
| 0.0 | 0.0 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.0 | GO:0032499 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0044393 | microspike(GO:0044393) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.0 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.0 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0099571 | postsynaptic actin cytoskeleton(GO:0098871) postsynaptic cytoskeleton(GO:0099571) |
| 0.0 | 0.1 | GO:0097227 | sperm annulus(GO:0097227) |
| 0.0 | 0.1 | GO:0031315 | extrinsic component of mitochondrial outer membrane(GO:0031315) |
| 0.0 | 0.2 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.0 | GO:0097648 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0031893 | vasopressin receptor binding(GO:0031893) |
| 0.1 | 0.3 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.1 | 0.2 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 1.4 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.1 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.2 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0031714 | C5a anaphylatoxin chemotactic receptor binding(GO:0031714) |
| 0.0 | 0.3 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.1 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.1 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.0 | 0.1 | GO:0008481 | sphinganine kinase activity(GO:0008481) |
| 0.0 | 0.1 | GO:0031871 | proteinase activated receptor binding(GO:0031871) |
| 0.0 | 0.1 | GO:0051990 | (R)-2-hydroxyglutarate dehydrogenase activity(GO:0051990) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.0 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.0 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.1 | GO:0046923 | ER retention sequence binding(GO:0046923) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.2 | GO:0005313 | L-glutamate transmembrane transporter activity(GO:0005313) |
| 0.0 | 0.0 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.1 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.0 | GO:0019136 | deoxynucleoside kinase activity(GO:0019136) |
| 0.0 | 0.0 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.0 | 0.0 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) nicotinate-nucleotide adenylyltransferase activity(GO:0004515) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.0 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.0 | REACTOME REGULATION OF GLUCOKINASE BY GLUCOKINASE REGULATORY PROTEIN | Genes involved in Regulation of Glucokinase by Glucokinase Regulatory Protein |
| 0.0 | 0.1 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |