Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
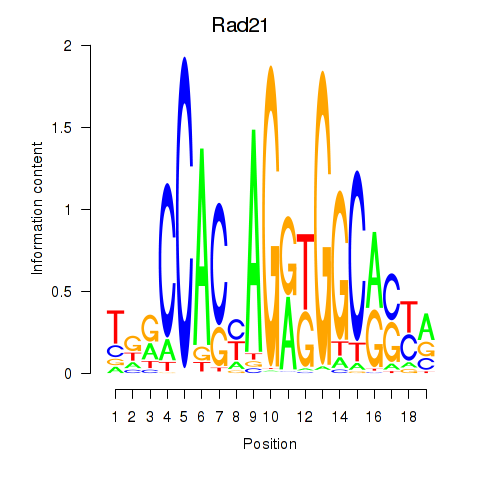
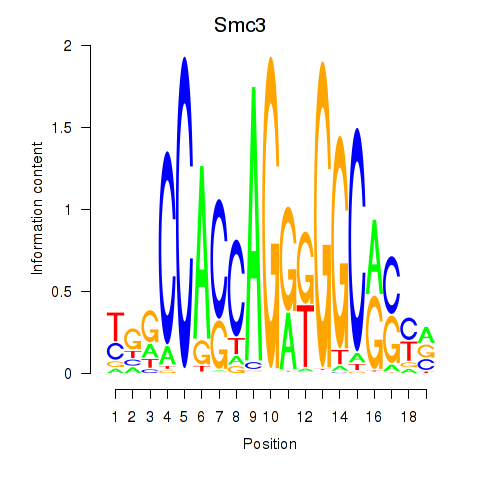
Results for Rad21_Smc3
Z-value: 0.76
Transcription factors associated with Rad21_Smc3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Rad21
|
ENSRNOG00000004420 | RAD21 cohesin complex component |
|
Smc3
|
ENSRNOG00000014173 | structural maintenance of chromosomes 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Smc3 | rn6_v1_chr1_+_274309758_274309758 | -0.65 | 2.3e-01 | Click! |
| Rad21 | rn6_v1_chr7_-_91538673_91538673 | -0.53 | 3.6e-01 | Click! |
Activity profile of Rad21_Smc3 motif
Sorted Z-values of Rad21_Smc3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_9695292 | 0.82 |
ENSRNOT00000036162
|
Prr7
|
proline rich 7 (synaptic) |
| chr3_+_164424515 | 0.81 |
ENSRNOT00000083876
|
Cebpb
|
CCAAT/enhancer binding protein beta |
| chr12_+_24651314 | 0.36 |
ENSRNOT00000077016
ENSRNOT00000071569 |
Vps37d
|
VPS37D, ESCRT-I subunit |
| chr16_+_19767264 | 0.35 |
ENSRNOT00000051802
ENSRNOT00000092073 |
Ocel1
|
occludin/ELL domain containing 1 |
| chr3_+_154395187 | 0.34 |
ENSRNOT00000050810
|
Vstm2l
|
V-set and transmembrane domain containing 2 like |
| chr19_+_41482728 | 0.34 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr13_+_75059927 | 0.33 |
ENSRNOT00000080801
|
LOC680254
|
hypothetical protein LOC680254 |
| chr8_+_71822129 | 0.32 |
ENSRNOT00000089147
|
Dapk2
|
death-associated protein kinase 2 |
| chrX_+_72684329 | 0.30 |
ENSRNOT00000057644
|
Dmrtc1c1
|
DMRT-like family C1c1 |
| chr16_+_20672374 | 0.29 |
ENSRNOT00000063780
|
RGD1566239
|
similar to RIKEN cDNA 2810428I15 |
| chr20_-_10680283 | 0.29 |
ENSRNOT00000001579
|
Sik1
|
salt-inducible kinase 1 |
| chr3_+_177374812 | 0.29 |
ENSRNOT00000024096
|
Polr3k
|
RNA polymerase III subunit K |
| chr12_+_48257609 | 0.29 |
ENSRNOT00000031658
ENSRNOT00000090277 |
Alkbh2
|
alkB homolog 2, alpha-ketoglutarate-dependent dioxygenase |
| chr10_-_85049331 | 0.27 |
ENSRNOT00000012538
|
Tbx21
|
T-box 21 |
| chr11_+_77815181 | 0.26 |
ENSRNOT00000002640
|
Cldn1
|
claudin 1 |
| chr15_-_29369504 | 0.26 |
ENSRNOT00000060297
|
AABR07017624.1
|
|
| chr15_-_34444244 | 0.26 |
ENSRNOT00000027612
|
Cideb
|
cell death-inducing DFFA-like effector b |
| chr17_-_13393243 | 0.25 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr20_+_3935529 | 0.25 |
ENSRNOT00000072458
|
LOC108348072
|
class II histocompatibility antigen, M alpha chain |
| chr1_+_79959591 | 0.24 |
ENSRNOT00000090515
|
Rsph6a
|
radial spoke head 6 homolog A |
| chr4_+_25435873 | 0.24 |
ENSRNOT00000000018
|
Steap1
|
STEAP family member 1 |
| chr10_-_13051154 | 0.24 |
ENSRNOT00000033457
|
AABR07029202.1
|
|
| chr1_+_82480195 | 0.23 |
ENSRNOT00000028051
|
Tgfb1
|
transforming growth factor, beta 1 |
| chr3_+_160231914 | 0.23 |
ENSRNOT00000014411
|
Kcnk15
|
potassium two pore domain channel subfamily K member 15 |
| chr9_-_43127887 | 0.23 |
ENSRNOT00000021685
|
Ankrd39
|
ankyrin repeat domain 39 |
| chr3_+_170252901 | 0.23 |
ENSRNOT00000005871
|
Mc3r
|
melanocortin 3 receptor |
| chr6_+_137997335 | 0.22 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr16_+_19874034 | 0.22 |
ENSRNOT00000023579
|
Mrpl34
|
mitochondrial ribosomal protein L34 |
| chr8_-_117237229 | 0.22 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr9_-_10441834 | 0.22 |
ENSRNOT00000043704
|
Rpl36
|
ribosomal protein L36 |
| chr7_+_140781799 | 0.21 |
ENSRNOT00000087932
|
Dnajc22
|
DnaJ heat shock protein family (Hsp40) member C22 |
| chr20_-_5244386 | 0.21 |
ENSRNOT00000070886
|
RT1-DMa
|
RT1 class II, locus DMa |
| chr3_-_3798177 | 0.21 |
ENSRNOT00000025386
ENSRNOT00000076156 |
Dnlz
|
DNL-type zinc finger |
| chr20_+_4593389 | 0.21 |
ENSRNOT00000001174
|
Slc44a4
|
solute carrier family 44, member 4 |
| chr15_-_23580342 | 0.20 |
ENSRNOT00000013331
ENSRNOT00000085767 |
Cnih1
|
cornichon family AMPA receptor auxiliary protein 1 |
| chr3_-_52998650 | 0.20 |
ENSRNOT00000009988
|
RGD1560831
|
similar to 40S ribosomal protein S3 |
| chr1_-_198706852 | 0.19 |
ENSRNOT00000024074
|
Dctpp1
|
dCTP pyrophosphatase 1 |
| chr1_-_242152834 | 0.19 |
ENSRNOT00000071614
ENSRNOT00000020412 |
Fxn
|
frataxin |
| chr11_+_88122271 | 0.19 |
ENSRNOT00000002540
|
Sdf2l1
|
stromal cell-derived factor 2-like 1 |
| chr3_-_154627257 | 0.19 |
ENSRNOT00000018328
|
Tgm2
|
transglutaminase 2 |
| chr5_+_143081326 | 0.19 |
ENSRNOT00000067204
|
Meaf6
|
MYST/Esa1-associated factor 6 |
| chr2_+_225827504 | 0.19 |
ENSRNOT00000018343
|
Gclm
|
glutamate cysteine ligase, modifier subunit |
| chr1_-_89258935 | 0.19 |
ENSRNOT00000003180
|
LOC100361079
|
ribosomal protein L36-like |
| chr11_+_33845463 | 0.19 |
ENSRNOT00000041838
|
Cbr1
|
carbonyl reductase 1 |
| chr4_+_62780596 | 0.18 |
ENSRNOT00000073846
|
LOC689574
|
hypothetical protein LOC689574 |
| chr4_-_171176581 | 0.17 |
ENSRNOT00000030850
|
Rerg
|
RAS-like, estrogen-regulated, growth-inhibitor |
| chr10_-_13187461 | 0.17 |
ENSRNOT00000007039
|
Prss41
|
protease, serine, 41 |
| chr3_-_72071895 | 0.17 |
ENSRNOT00000082857
|
Selenoh
|
selenoprotein H |
| chr5_+_122508388 | 0.17 |
ENSRNOT00000038410
|
Tctex1d1
|
Tctex1 domain containing 1 |
| chrX_+_45637415 | 0.17 |
ENSRNOT00000050544
|
RGD1563378
|
similar to ferritin heavy polypeptide-like 17 |
| chr11_-_67756799 | 0.17 |
ENSRNOT00000030975
|
Parp9
|
poly (ADP-ribose) polymerase family, member 9 |
| chr15_-_38276159 | 0.16 |
ENSRNOT00000084698
ENSRNOT00000015233 |
Micu2
|
mitochondrial calcium uptake 2 |
| chr16_+_9907598 | 0.16 |
ENSRNOT00000027352
|
Ptpn20
|
protein tyrosine phosphatase, non-receptor type 20 |
| chr16_+_21282467 | 0.16 |
ENSRNOT00000065345
|
Yjefn3
|
YjeF N-terminal domain containing 3 |
| chr10_+_55492404 | 0.16 |
ENSRNOT00000005588
ENSRNOT00000078038 |
Rpl26
|
ribosomal protein L26 |
| chr5_-_151397603 | 0.16 |
ENSRNOT00000076866
|
Gpr3
|
G protein-coupled receptor 3 |
| chr1_+_91363492 | 0.16 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr15_+_32386816 | 0.15 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr2_+_200397967 | 0.15 |
ENSRNOT00000025821
|
Reg4
|
regenerating family member 4 |
| chr7_+_139271698 | 0.15 |
ENSRNOT00000079388
|
Slc48a1
|
solute carrier family 48 member 1 |
| chr5_-_164927869 | 0.15 |
ENSRNOT00000012080
|
Draxin
|
dorsal inhibitory axon guidance protein |
| chr3_+_148150698 | 0.15 |
ENSRNOT00000087075
ENSRNOT00000010251 |
Hm13
|
histocompatibility minor 13 |
| chr2_+_190007216 | 0.15 |
ENSRNOT00000015612
|
S100a6
|
S100 calcium binding protein A6 |
| chr13_-_110257367 | 0.15 |
ENSRNOT00000005576
|
Dtl
|
denticleless E3 ubiquitin protein ligase homolog |
| chr9_-_92775816 | 0.15 |
ENSRNOT00000029635
|
RGD1563917
|
similar to Nuclear autoantigen Sp-100 (Speckled 100 kDa) |
| chr2_-_219558675 | 0.14 |
ENSRNOT00000020149
|
Rtcd1
|
RNA terminal phosphate cyclase domain 1 |
| chr3_+_175548174 | 0.14 |
ENSRNOT00000091941
|
Adrm1
|
adhesion regulating molecule 1 |
| chr20_+_6869767 | 0.14 |
ENSRNOT00000000631
ENSRNOT00000086330 ENSRNOT00000093188 ENSRNOT00000093736 |
RGD735065
|
similar to GI:13385412-like protein splice form I |
| chr2_+_86951776 | 0.14 |
ENSRNOT00000087275
|
AABR07009105.1
|
|
| chr2_-_199771896 | 0.14 |
ENSRNOT00000043937
|
Chd1l
|
chromodomain helicase DNA binding protein 1-like |
| chr2_+_251817694 | 0.14 |
ENSRNOT00000019964
|
RGD1560065
|
similar to RIKEN cDNA 2410004B18 |
| chr20_-_3374344 | 0.14 |
ENSRNOT00000082999
|
Ppp1r18
|
protein phosphatase 1, regulatory subunit 18 |
| chr17_+_10384511 | 0.14 |
ENSRNOT00000024357
|
Sncb
|
synuclein, beta |
| chrX_-_72078551 | 0.13 |
ENSRNOT00000076978
|
Rps4x
|
ribosomal protein S4, X-linked |
| chr1_-_80666566 | 0.13 |
ENSRNOT00000082125
ENSRNOT00000025388 |
Nectin2
|
nectin cell adhesion molecule 2 |
| chr6_-_11494459 | 0.13 |
ENSRNOT00000021570
|
Kcnk12
|
potassium two pore domain channel subfamily K member 12 |
| chr7_-_138707221 | 0.13 |
ENSRNOT00000009199
|
Amigo2
|
adhesion molecule with Ig like domain 2 |
| chr17_+_57075218 | 0.13 |
ENSRNOT00000089536
|
Crem
|
cAMP responsive element modulator |
| chr10_+_55013703 | 0.13 |
ENSRNOT00000032785
|
Pik3r5
|
phosphoinositide-3-kinase, regulatory subunit 5 |
| chr1_-_226292480 | 0.13 |
ENSRNOT00000039133
|
Myrf
|
myelin regulatory factor |
| chr3_+_56966654 | 0.13 |
ENSRNOT00000088097
|
Gorasp2
|
golgi reassembly stacking protein 2 |
| chr10_+_105861743 | 0.13 |
ENSRNOT00000064410
|
Mgat5b
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B |
| chrX_-_131343853 | 0.13 |
ENSRNOT00000038653
|
Dcaf12l1
|
DDB1 and CUL4 associated factor 12-like 1 |
| chr5_-_61077627 | 0.13 |
ENSRNOT00000015338
|
Shb
|
SH2 domain containing adaptor protein B |
| chr7_-_2909144 | 0.12 |
ENSRNOT00000082518
ENSRNOT00000089074 ENSRNOT00000085644 |
Myl6
|
myosin light chain 6 |
| chr19_+_37652969 | 0.12 |
ENSRNOT00000041970
|
Carmil2
|
capping protein regulator and myosin 1 linker 2 |
| chr8_-_110813000 | 0.12 |
ENSRNOT00000010634
|
Ephb1
|
Eph receptor B1 |
| chr12_-_46794797 | 0.12 |
ENSRNOT00000001518
|
Rplp0
|
ribosomal protein lateral stalk subunit P0 |
| chr1_-_52962388 | 0.12 |
ENSRNOT00000033685
|
T2
|
brachyury 2 |
| chr6_+_136720582 | 0.12 |
ENSRNOT00000086868
|
Kif26a
|
kinesin family member 26A |
| chr3_+_160908769 | 0.12 |
ENSRNOT00000030054
|
Sys1
|
Sys1 golgi trafficking protein |
| chr5_-_60658521 | 0.12 |
ENSRNOT00000092866
ENSRNOT00000093024 ENSRNOT00000016875 |
Tomm5
Fbxo10
|
translocase of outer mitochondrial membrane 5 F-box protein 10 |
| chr16_+_9563218 | 0.11 |
ENSRNOT00000035915
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr3_+_14588870 | 0.11 |
ENSRNOT00000071712
|
LOC100912604
|
spermidine synthase-like |
| chr4_+_170958196 | 0.11 |
ENSRNOT00000007905
|
Pde6h
|
phosphodiesterase 6H |
| chr8_-_104912959 | 0.11 |
ENSRNOT00000017363
|
Spsb4
|
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr10_-_46332172 | 0.11 |
ENSRNOT00000004475
|
Rasd1
|
ras related dexamethasone induced 1 |
| chr5_-_159602251 | 0.11 |
ENSRNOT00000011394
|
Necap2
|
NECAP endocytosis associated 2 |
| chr9_+_98505259 | 0.11 |
ENSRNOT00000051810
|
Erfe
|
erythroferrone |
| chr15_+_31948035 | 0.11 |
ENSRNOT00000071627
|
AABR07017868.1
|
|
| chr15_+_2631529 | 0.11 |
ENSRNOT00000018897
|
Comtd1
|
catechol-O-methyltransferase domain containing 1 |
| chr10_+_13723405 | 0.11 |
ENSRNOT00000072789
|
Abca3
|
ATP binding cassette subfamily A member 3 |
| chr5_-_156141537 | 0.11 |
ENSRNOT00000019004
|
Alpl
|
alkaline phosphatase, liver/bone/kidney |
| chr8_-_130491998 | 0.11 |
ENSRNOT00000064114
|
Higd1a
|
HIG1 hypoxia inducible domain family, member 1A |
| chr9_-_15306465 | 0.11 |
ENSRNOT00000019404
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chr19_-_15733412 | 0.10 |
ENSRNOT00000014831
|
Irx6
|
iroquois homeobox 6 |
| chr4_+_155313671 | 0.10 |
ENSRNOT00000020812
|
Mfap5
|
microfibrillar associated protein 5 |
| chr19_+_38397466 | 0.10 |
ENSRNOT00000034386
|
Nob1
|
NIN1/PSMD8 binding protein 1 homolog |
| chr3_+_176093640 | 0.10 |
ENSRNOT00000086541
|
Ogfr
|
opioid growth factor receptor |
| chr6_+_136720266 | 0.10 |
ENSRNOT00000018278
|
Kif26a
|
kinesin family member 26A |
| chr5_+_169506138 | 0.10 |
ENSRNOT00000014904
|
Rpl22
|
ribosomal protein L22 |
| chr19_-_10620671 | 0.10 |
ENSRNOT00000021842
|
Ccl17
|
C-C motif chemokine ligand 17 |
| chr1_-_85517360 | 0.10 |
ENSRNOT00000026114
|
Eid2
|
EP300 interacting inhibitor of differentiation 2 |
| chr17_+_26785029 | 0.10 |
ENSRNOT00000022065
|
Eef1e1
|
eukaryotic translation elongation factor 1 epsilon 1 |
| chr2_+_238529074 | 0.10 |
ENSRNOT00000016195
|
Ppa2
|
pyrophosphatase (inorganic) 2 |
| chr1_+_101213788 | 0.10 |
ENSRNOT00000090838
|
Tead2
|
TEA domain transcription factor 2 |
| chr10_-_14092289 | 0.10 |
ENSRNOT00000019624
|
Ndufb10
|
NADH:ubiquinone oxidoreductase subunit B10 |
| chr16_+_81204766 | 0.09 |
ENSRNOT00000042465
|
Tmem255b
|
transmembrane protein 255B |
| chr5_+_104941066 | 0.09 |
ENSRNOT00000009225
|
Rraga
|
Ras-related GTP binding A |
| chr15_+_12407524 | 0.09 |
ENSRNOT00000009249
|
Psmd6
|
proteasome 26S subunit, non-ATPase 6 |
| chr3_+_168124673 | 0.09 |
ENSRNOT00000070809
|
Pfdn4
|
prefoldin subunit 4 |
| chr13_-_50509916 | 0.09 |
ENSRNOT00000076747
|
Ren
|
renin |
| chr20_+_7818289 | 0.09 |
ENSRNOT00000042539
|
Ppard
|
peroxisome proliferator-activated receptor delta |
| chr3_+_72191533 | 0.09 |
ENSRNOT00000044319
|
Ube2l6
|
ubiquitin-conjugating enzyme E2L 6 |
| chr11_+_36851038 | 0.09 |
ENSRNOT00000002221
ENSRNOT00000061047 |
Pcp4
|
Purkinje cell protein 4 |
| chr13_+_79886832 | 0.09 |
ENSRNOT00000077069
ENSRNOT00000035558 ENSRNOT00000076417 |
Pigc
|
phosphatidylinositol glycan anchor biosynthesis, class C |
| chr11_-_72109964 | 0.09 |
ENSRNOT00000058917
|
AABR07034445.1
|
|
| chr6_-_128989812 | 0.09 |
ENSRNOT00000085943
|
LOC100909439
|
ankyrin repeat and SOCS box protein 2-like |
| chr5_-_2632030 | 0.09 |
ENSRNOT00000009096
|
Rdh10
|
retinol dehydrogenase 10 |
| chr19_-_37952501 | 0.09 |
ENSRNOT00000026809
|
Dpep3
|
dipeptidase 3 |
| chrX_-_105390580 | 0.09 |
ENSRNOT00000077547
|
Btk
|
Bruton tyrosine kinase |
| chr5_-_107857320 | 0.09 |
ENSRNOT00000008898
|
Cdkn2b
|
cyclin-dependent kinase inhibitor 2B |
| chr11_+_86516390 | 0.09 |
ENSRNOT00000041168
|
Gp1bb
|
glycoprotein Ib platelet beta subunit |
| chr10_-_97582188 | 0.09 |
ENSRNOT00000005076
|
Rgs9
|
regulator of G-protein signaling 9 |
| chr19_-_26243223 | 0.09 |
ENSRNOT00000037766
|
Zfp791
|
zinc finger protein 791 |
| chr10_-_57618527 | 0.09 |
ENSRNOT00000037517
|
C1qbp
|
complement C1q binding protein |
| chr11_+_73738433 | 0.08 |
ENSRNOT00000002353
|
Tmem44
|
transmembrane protein 44 |
| chr3_+_164274710 | 0.08 |
ENSRNOT00000012939
|
Snai1
|
snail family transcriptional repressor 1 |
| chr1_-_216920625 | 0.08 |
ENSRNOT00000028119
|
Osbpl5
|
oxysterol binding protein-like 5 |
| chr14_+_86029335 | 0.08 |
ENSRNOT00000017375
|
Dbnl
|
drebrin-like |
| chr1_+_173532803 | 0.08 |
ENSRNOT00000021017
|
Eif3f
|
eukaryotic translation initiation factor 3, subunit F |
| chr1_-_101131012 | 0.08 |
ENSRNOT00000082283
ENSRNOT00000093498 ENSRNOT00000093559 |
Flt3lg
Rpl13a
|
fms-related tyrosine kinase 3 ligand ribosomal protein L13A |
| chr12_+_13299859 | 0.08 |
ENSRNOT00000039191
|
LOC683674
|
similar to Protein C7orf26 homolog |
| chr8_+_128806129 | 0.08 |
ENSRNOT00000025225
|
Rpsa
|
ribosomal protein SA |
| chr2_+_207271853 | 0.08 |
ENSRNOT00000085355
ENSRNOT00000017254 |
Rhoc
|
ras homolog family member C |
| chr10_-_70241254 | 0.08 |
ENSRNOT00000038376
|
Rad51d
|
RAD51 paralog D |
| chr2_+_198772937 | 0.08 |
ENSRNOT00000028812
|
Itga10
|
integrin subunit alpha 10 |
| chr14_+_83752393 | 0.08 |
ENSRNOT00000081123
|
Selenom
|
selenoprotein M |
| chr13_+_96195836 | 0.08 |
ENSRNOT00000042547
|
RGD1563812
|
similar to basic transcription factor 3 |
| chr1_+_221773254 | 0.08 |
ENSRNOT00000028646
|
Rasgrp2
|
RAS guanyl releasing protein 2 |
| chr4_+_117589591 | 0.08 |
ENSRNOT00000078239
|
LOC103690067
|
EKC/KEOPS complex subunit Tprkb |
| chr10_-_19715832 | 0.08 |
ENSRNOT00000040278
|
RGD1564698
|
similar to ribosomal protein S10 |
| chr1_-_190598122 | 0.08 |
ENSRNOT00000044384
|
Pdzd9
|
PDZ domain containing 9 |
| chr1_-_89483988 | 0.08 |
ENSRNOT00000028603
|
Fxyd7
|
FXYD domain-containing ion transport regulator 7 |
| chr1_-_8878136 | 0.08 |
ENSRNOT00000064836
ENSRNOT00000075850 |
Vta1
|
vesicle trafficking 1 |
| chrX_+_70322755 | 0.08 |
ENSRNOT00000057900
|
Igbp1
|
immunoglobulin (CD79A) binding protein 1 |
| chr14_+_10581136 | 0.08 |
ENSRNOT00000002987
|
Coq2
|
coenzyme Q2, polyprenyltransferase |
| chr7_-_11699436 | 0.08 |
ENSRNOT00000049221
ENSRNOT00000066037 |
Tmprss9
|
transmembrane protease, serine 9 |
| chr5_+_173274774 | 0.08 |
ENSRNOT00000025952
|
Ccnl2
|
cyclin L2 |
| chr1_-_198298076 | 0.08 |
ENSRNOT00000027033
|
Ino80e
|
INO80 complex subunit E |
| chr13_-_90832138 | 0.08 |
ENSRNOT00000010930
|
Igsf9
|
immunoglobulin superfamily, member 9 |
| chr18_-_37776453 | 0.07 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr1_-_52354175 | 0.07 |
ENSRNOT00000015010
|
MGC94891
|
hypothetical protein LOC681210 |
| chr1_-_1116926 | 0.07 |
ENSRNOT00000062027
|
Raet1l
|
retinoic acid early transcript 1L |
| chr7_-_12793711 | 0.07 |
ENSRNOT00000013762
|
Palm
|
paralemmin |
| chr1_+_221792221 | 0.07 |
ENSRNOT00000054828
|
Nrxn2
|
neurexin 2 |
| chr10_-_109757550 | 0.07 |
ENSRNOT00000054957
|
Arhgdia
|
Rho GDP dissociation inhibitor alpha |
| chr18_-_27206777 | 0.07 |
ENSRNOT00000090569
|
AABR07031691.1
|
|
| chr1_+_220243473 | 0.07 |
ENSRNOT00000084650
ENSRNOT00000027091 |
mrpl11
|
mitochondrial ribosomal protein L11 |
| chr3_+_146980923 | 0.07 |
ENSRNOT00000011654
|
Nsfl1c
|
NSFL1 cofactor |
| chr6_+_75429729 | 0.07 |
ENSRNOT00000043250
|
Rps10l1
|
ribosomal protein S10-like 1 |
| chr2_-_198719202 | 0.07 |
ENSRNOT00000028801
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr1_-_101131413 | 0.07 |
ENSRNOT00000093729
|
Flt3lg
|
fms-related tyrosine kinase 3 ligand |
| chr9_+_100076958 | 0.07 |
ENSRNOT00000084297
|
Dusp28
|
dual specificity phosphatase 28 |
| chrX_-_1741701 | 0.07 |
ENSRNOT00000031115
|
Uba1
|
ubiquitin-like modifier activating enzyme 1 |
| chr12_+_22165486 | 0.07 |
ENSRNOT00000001890
|
Mospd3
|
motile sperm domain containing 3 |
| chr13_-_68360664 | 0.07 |
ENSRNOT00000030971
|
Hmcn1
|
hemicentin 1 |
| chr8_-_62424303 | 0.07 |
ENSRNOT00000091223
|
Csk
|
c-src tyrosine kinase |
| chr11_-_72157522 | 0.07 |
ENSRNOT00000002373
|
Meltf
|
melanotransferrin |
| chr13_-_112005052 | 0.07 |
ENSRNOT00000007879
|
G0s2
|
G0/G1switch 2 |
| chr17_+_9631925 | 0.07 |
ENSRNOT00000018114
|
Ddx41
|
DEAD-box helicase 41 |
| chr2_+_248249468 | 0.06 |
ENSRNOT00000022648
|
Gbp4
|
guanylate binding protein 4 |
| chr7_+_12006710 | 0.06 |
ENSRNOT00000045421
|
Klf16
|
Kruppel-like factor 16 |
| chr14_+_78213635 | 0.06 |
ENSRNOT00000010541
|
Evc
|
EvC ciliary complex subunit 1 |
| chr12_-_30566032 | 0.06 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr9_+_10941613 | 0.06 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr20_-_32416044 | 0.06 |
ENSRNOT00000067004
|
Ccar1
|
cell division cycle and apoptosis regulator 1 |
| chr19_+_60104804 | 0.06 |
ENSRNOT00000078559
|
Pard3
|
par-3 family cell polarity regulator |
| chr1_-_101426852 | 0.06 |
ENSRNOT00000028217
|
Ruvbl2
|
RuvB-like AAA ATPase 2 |
| chr10_+_103703404 | 0.06 |
ENSRNOT00000086469
|
Rab37
|
RAB37, member RAS oncogene family |
| chrX_-_115496045 | 0.06 |
ENSRNOT00000051134
|
LOC100361854
|
ribosomal protein S26-like |
| chr2_-_188471988 | 0.06 |
ENSRNOT00000027785
|
Hcn3
|
hyperpolarization-activated cyclic nucleotide-gated potassium channel 3 |
| chr1_+_78818404 | 0.06 |
ENSRNOT00000090417
|
Gng8
|
G protein subunit gamma 8 |
| chr20_+_7908304 | 0.06 |
ENSRNOT00000000603
|
Rpl10a
|
ribosomal protein L10A |
| chr2_-_44907030 | 0.06 |
ENSRNOT00000013979
|
Gpx8
|
glutathione peroxidase 8 |
| chr10_+_56260514 | 0.06 |
ENSRNOT00000016177
|
Sox15
|
SRY box 15 |
| chr2_-_185303610 | 0.06 |
ENSRNOT00000093479
ENSRNOT00000046286 |
NEWGENE_1592020
|
protease, serine 48 |
| chr15_+_31462806 | 0.06 |
ENSRNOT00000072946
|
AABR07017825.3
|
|
| chr9_+_40975836 | 0.06 |
ENSRNOT00000084470
|
Ptpn18
|
protein tyrosine phosphatase, non-receptor type 18 |
| chr20_-_14282873 | 0.06 |
ENSRNOT00000001759
|
Adora2a
|
adenosine A2a receptor |
Network of associatons between targets according to the STRING database.
First level regulatory network of Rad21_Smc3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0035711 | granuloma formation(GO:0002432) T-helper 1 cell activation(GO:0035711) |
| 0.1 | 0.3 | GO:2000422 | regulation of eosinophil chemotaxis(GO:2000422) positive regulation of eosinophil chemotaxis(GO:2000424) |
| 0.1 | 0.3 | GO:1903544 | positive regulation of bicellular tight junction assembly(GO:1903348) response to butyrate(GO:1903544) |
| 0.1 | 0.2 | GO:1900126 | negative regulation of hyaluronan biosynthetic process(GO:1900126) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.2 | GO:0018283 | metal incorporation into metallo-sulfur cluster(GO:0018282) iron incorporation into metallo-sulfur cluster(GO:0018283) |
| 0.1 | 0.2 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.1 | 0.2 | GO:0042374 | phylloquinone metabolic process(GO:0042374) phylloquinone catabolic process(GO:0042376) quinone catabolic process(GO:1901662) |
| 0.1 | 0.3 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.1 | 0.2 | GO:1902167 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage by p53 class mediator(GO:1902167) |
| 0.1 | 0.2 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) |
| 0.0 | 0.1 | GO:1904211 | membrane protein proteolysis involved in retrograde protein transport, ER to cytosol(GO:1904211) |
| 0.0 | 0.2 | GO:0009223 | pyrimidine deoxyribonucleotide catabolic process(GO:0009223) |
| 0.0 | 0.1 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.0 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.2 | GO:0008292 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.0 | GO:0002458 | peripheral T cell tolerance induction(GO:0002458) peripheral tolerance induction(GO:0002465) |
| 0.0 | 0.3 | GO:0032792 | negative regulation of CREB transcription factor activity(GO:0032792) |
| 0.0 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.1 | GO:0071529 | cementum mineralization(GO:0071529) |
| 0.0 | 0.2 | GO:0002503 | MHC class II protein complex assembly(GO:0002399) peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) |
| 0.0 | 0.1 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 | 0.1 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.1 | GO:0006407 | rRNA export from nucleus(GO:0006407) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:1904401 | cellular response to Thyroid stimulating hormone(GO:1904401) |
| 0.0 | 0.2 | GO:0097577 | intracellular sequestering of iron ion(GO:0006880) sequestering of iron ion(GO:0097577) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0008295 | spermidine biosynthetic process(GO:0008295) |
| 0.0 | 0.1 | GO:1904009 | cellular response to monosodium glutamate(GO:1904009) |
| 0.0 | 0.1 | GO:0071226 | positive regulation of type I hypersensitivity(GO:0001812) peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.1 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:0090579 | transcriptional activation by promoter-enhancer looping(GO:0071733) regulation of estrogen receptor binding(GO:0071898) negative regulation of estrogen receptor binding(GO:0071899) gene looping(GO:0090202) dsDNA loop formation(GO:0090579) |
| 0.0 | 0.1 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.0 | 0.0 | GO:0070343 | white fat cell proliferation(GO:0070343) regulation of white fat cell proliferation(GO:0070350) |
| 0.0 | 0.2 | GO:0044154 | histone H3-K14 acetylation(GO:0044154) |
| 0.0 | 0.1 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0060160 | negative regulation of dopamine receptor signaling pathway(GO:0060160) |
| 0.0 | 0.1 | GO:0002018 | angiotensin maturation(GO:0002003) renin-angiotensin regulation of aldosterone production(GO:0002018) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0060431 | primary lung bud formation(GO:0060431) |
| 0.0 | 0.2 | GO:0000050 | urea cycle(GO:0000050) |
| 0.0 | 0.2 | GO:0015886 | heme transport(GO:0015886) |
| 0.0 | 0.2 | GO:0071712 | ER-associated misfolded protein catabolic process(GO:0071712) |
| 0.0 | 0.0 | GO:1902071 | positive regulation of cellular response to hypoxia(GO:1900039) regulation of hypoxia-inducible factor-1alpha signaling pathway(GO:1902071) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.0 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.0 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.0 | 0.1 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.1 | GO:0014057 | positive regulation of acetylcholine secretion, neurotransmission(GO:0014057) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.2 | GO:0019985 | translesion synthesis(GO:0019985) |
| 0.0 | 0.2 | GO:0060330 | regulation of response to interferon-gamma(GO:0060330) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.2 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.2 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:2000774 | positive regulation of cellular senescence(GO:2000774) |
| 0.0 | 0.1 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.2 | GO:0002504 | antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002504) |
| 0.0 | 0.0 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) |
| 0.0 | 0.1 | GO:0090258 | negative regulation of mitochondrial fission(GO:0090258) |
| 0.0 | 0.0 | GO:1904158 | axonemal central apparatus assembly(GO:1904158) |
| 0.0 | 0.1 | GO:0021631 | optic nerve morphogenesis(GO:0021631) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0036488 | CHOP-C/EBP complex(GO:0036488) |
| 0.0 | 0.1 | GO:0005944 | phosphatidylinositol 3-kinase complex, class IB(GO:0005944) |
| 0.0 | 0.4 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0097233 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.4 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.1 | GO:0071556 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) integral component of lumenal side of endoplasmic reticulum membrane(GO:0071556) lumenal side of endoplasmic reticulum membrane(GO:0098553) |
| 0.0 | 0.1 | GO:1990730 | VCP-NSFL1C complex(GO:1990730) |
| 0.0 | 0.2 | GO:0065010 | extracellular membrane-bounded organelle(GO:0065010) |
| 0.0 | 0.2 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.2 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.2 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.1 | GO:0045298 | tubulin complex(GO:0045298) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.1 | GO:0008290 | F-actin capping protein complex(GO:0008290) |
| 0.0 | 0.1 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.1 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.1 | GO:0044316 | cone cell pedicle(GO:0044316) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.3 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0016282 | eukaryotic 43S preinitiation complex(GO:0016282) eukaryotic 48S preinitiation complex(GO:0033290) eukaryotic translation initiation factor 3 complex, eIF3m(GO:0071541) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.1 | 0.2 | GO:0004977 | melanocortin receptor activity(GO:0004977) |
| 0.0 | 0.2 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.2 | GO:0004090 | carbonyl reductase (NADPH) activity(GO:0004090) |
| 0.0 | 0.2 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.2 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.3 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.7 | GO:0035259 | glucocorticoid receptor binding(GO:0035259) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0008158 | hedgehog receptor activity(GO:0008158) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.2 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.1 | GO:0004427 | inorganic diphosphatase activity(GO:0004427) |
| 0.0 | 0.2 | GO:0047429 | deoxyribonucleotide binding(GO:0032552) nucleoside-triphosphate diphosphatase activity(GO:0047429) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.1 | GO:0031750 | D3 dopamine receptor binding(GO:0031750) |
| 0.0 | 0.1 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0001013 | RNA polymerase I regulatory region DNA binding(GO:0001013) |
| 0.0 | 0.1 | GO:0099609 | microtubule lateral binding(GO:0099609) |
| 0.0 | 0.2 | GO:0015232 | heme transporter activity(GO:0015232) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0016886 | ligase activity, forming phosphoric ester bonds(GO:0016886) |
| 0.0 | 0.1 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.0 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0015189 | L-lysine transmembrane transporter activity(GO:0015189) |
| 0.0 | 0.1 | GO:0004035 | alkaline phosphatase activity(GO:0004035) |
| 0.0 | 0.1 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.0 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.0 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.0 | 0.1 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.2 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.1 | GO:0043141 | ATP-dependent 5'-3' DNA helicase activity(GO:0043141) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID IL3 PATHWAY | IL3-mediated signaling events |
| 0.0 | 0.5 | PID IL27 PATHWAY | IL27-mediated signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.3 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |