Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
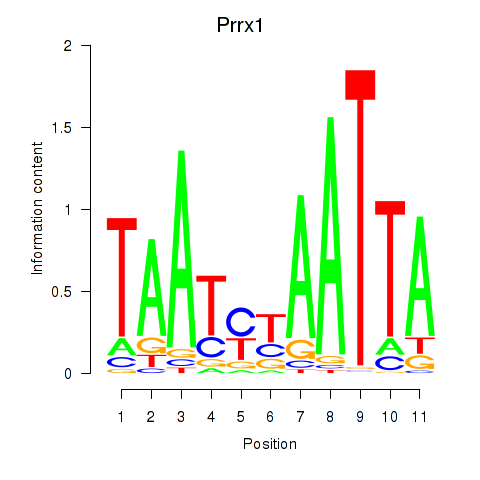
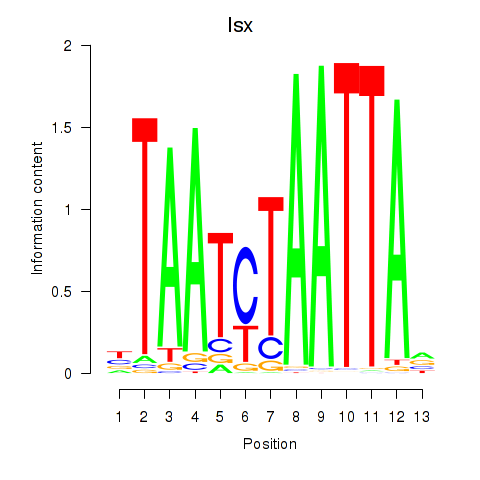
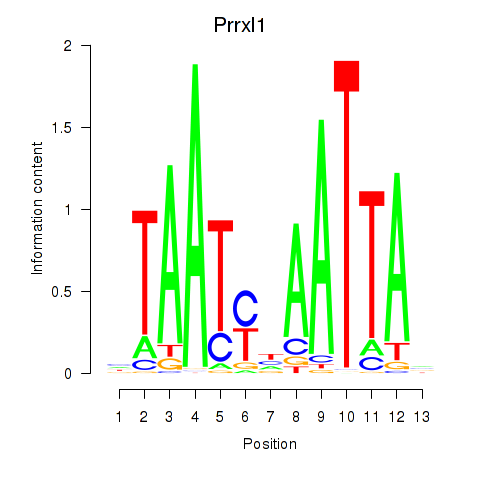
Results for Prrx1_Isx_Prrxl1
Z-value: 0.48
Transcription factors associated with Prrx1_Isx_Prrxl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prrx1
|
ENSRNOG00000003720 | paired related homeobox 1 |
|
Isx
|
ENSRNOG00000039808 | intestine-specific homeobox |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prrx1 | rn6_v1_chr13_-_81214494_81214546 | 0.55 | 3.4e-01 | Click! |
| Drgx | rn6_v1_chr16_+_8823872_8823872 | 0.35 | 5.7e-01 | Click! |
Activity profile of Prrx1_Isx_Prrxl1 motif
Sorted Z-values of Prrx1_Isx_Prrxl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_47677720 | 0.32 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr14_+_69800156 | 0.29 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr8_-_102149912 | 0.28 |
ENSRNOT00000011263
|
RGD1309079
|
similar to Ab2-095 |
| chr3_+_155160481 | 0.26 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr4_+_51614676 | 0.23 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr18_-_32207749 | 0.23 |
ENSRNOT00000090882
ENSRNOT00000018843 |
Arhgap26
|
Rho GTPase activating protein 26 |
| chr2_+_198303168 | 0.22 |
ENSRNOT00000056262
|
Mtmr11
|
myotubularin related protein 11 |
| chr10_+_56381813 | 0.22 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr1_-_167911961 | 0.21 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr13_-_55561726 | 0.21 |
ENSRNOT00000000817
ENSRNOT00000090162 |
Nek7
|
NIMA-related kinase 7 |
| chr7_-_101138860 | 0.19 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr7_-_52404774 | 0.18 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr8_-_39551700 | 0.18 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr13_+_91054974 | 0.17 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr2_-_117454769 | 0.17 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr2_-_38110567 | 0.16 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chr2_-_149444548 | 0.16 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr8_+_82038967 | 0.16 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr2_+_4989295 | 0.16 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
| chr15_+_62406873 | 0.15 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr17_+_63635086 | 0.15 |
ENSRNOT00000020634
|
Dip2c
|
disco-interacting protein 2 homolog C |
| chr8_-_78397123 | 0.15 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr4_-_155740193 | 0.14 |
ENSRNOT00000043229
|
LOC102550396
|
LRRGT00188 |
| chr16_+_32457521 | 0.14 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr4_-_55011415 | 0.12 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr18_+_27558089 | 0.12 |
ENSRNOT00000027499
|
Fam53c
|
family with sequence similarity 53, member C |
| chr8_-_132753145 | 0.11 |
ENSRNOT00000007467
ENSRNOT00000008172 |
RGD1566368
|
similar to Solute carrier family 6 (neurotransmitter transporter), member 20 |
| chr5_+_118574801 | 0.11 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr1_-_245817809 | 0.11 |
ENSRNOT00000080607
|
AABR07006672.1
|
|
| chr20_+_6211420 | 0.11 |
ENSRNOT00000000624
|
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr2_+_148262110 | 0.11 |
ENSRNOT00000046641
|
AABR07010705.1
|
|
| chr7_+_141666111 | 0.11 |
ENSRNOT00000083250
|
AABR07058886.1
|
|
| chr7_+_27174882 | 0.10 |
ENSRNOT00000051181
|
Glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr1_-_88162583 | 0.10 |
ENSRNOT00000087411
|
Catsperg
|
cation channel sperm associated auxiliary subunit gamma |
| chr10_+_65552897 | 0.10 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr7_-_107768072 | 0.10 |
ENSRNOT00000093189
|
Ndrg1
|
N-myc downstream regulated 1 |
| chr10_-_16731898 | 0.09 |
ENSRNOT00000028186
|
Crebrf
|
CREB3 regulatory factor |
| chr18_+_30398113 | 0.09 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr1_-_67134827 | 0.09 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr3_+_81134505 | 0.09 |
ENSRNOT00000067664
|
Phf21a
|
PHD finger protein 21A |
| chr18_-_32670665 | 0.09 |
ENSRNOT00000019409
|
Nr3c1
|
nuclear receptor subfamily 3, group C, member 1 |
| chr2_+_196608496 | 0.09 |
ENSRNOT00000091681
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr4_+_70252366 | 0.09 |
ENSRNOT00000073039
|
Chl1
|
cell adhesion molecule L1-like |
| chr13_-_76049363 | 0.09 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chrX_+_104882704 | 0.09 |
ENSRNOT00000079572
ENSRNOT00000074330 ENSRNOT00000082983 |
Cstf2
|
cleavage stimulation factor subunit 2 |
| chr2_+_86891092 | 0.09 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr18_+_16146447 | 0.09 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr15_+_56666012 | 0.09 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr11_+_34865532 | 0.08 |
ENSRNOT00000050342
|
Dyrk1a
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr1_+_234363994 | 0.08 |
ENSRNOT00000018137
|
Rorb
|
RAR-related orphan receptor B |
| chr2_-_86475096 | 0.08 |
ENSRNOT00000075247
ENSRNOT00000075062 |
LOC100909688
|
zinc finger protein 43-like |
| chr9_+_111597037 | 0.08 |
ENSRNOT00000021758
|
Fer
|
FER tyrosine kinase |
| chr16_+_78539489 | 0.08 |
ENSRNOT00000057897
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr15_-_88670349 | 0.08 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr7_-_104801045 | 0.08 |
ENSRNOT00000079524
|
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr5_+_135574172 | 0.08 |
ENSRNOT00000023416
|
Tesk2
|
testis-specific kinase 2 |
| chr11_-_72814850 | 0.08 |
ENSRNOT00000091575
|
LOC100911374
|
UBX domain-containing protein 7-like |
| chr14_+_39663421 | 0.07 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr12_-_47990105 | 0.07 |
ENSRNOT00000080049
|
Ube3b
|
ubiquitin protein ligase E3B |
| chr15_+_4209703 | 0.07 |
ENSRNOT00000082236
|
Ppp3cb
|
protein phosphatase 3 catalytic subunit beta |
| chr5_+_156439633 | 0.07 |
ENSRNOT00000090232
|
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr7_+_117456039 | 0.07 |
ENSRNOT00000051754
|
Mroh1
|
maestro heat-like repeat family member 1 |
| chr17_+_55065621 | 0.07 |
ENSRNOT00000086342
|
Zfp438
|
zinc finger protein 438 |
| chr2_+_266315036 | 0.07 |
ENSRNOT00000055245
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr3_-_138683318 | 0.07 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr10_-_87286387 | 0.07 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr1_-_53152866 | 0.07 |
ENSRNOT00000085501
|
Fgfr1op
|
Fgfr1 oncogene partner |
| chr16_+_84465656 | 0.07 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr10_+_66942398 | 0.07 |
ENSRNOT00000018986
|
Rab11fip4
|
RAB11 family interacting protein 4 |
| chr2_-_40386669 | 0.07 |
ENSRNOT00000014074
|
Elovl7
|
ELOVL fatty acid elongase 7 |
| chr2_+_86996497 | 0.07 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr8_-_69508024 | 0.07 |
ENSRNOT00000083564
|
AABR07070416.3
|
|
| chr7_-_120770435 | 0.07 |
ENSRNOT00000077000
|
Ddx17
|
DEAD-box helicase 17 |
| chr19_+_25043680 | 0.06 |
ENSRNOT00000043971
|
Adgrl1
|
adhesion G protein-coupled receptor L1 |
| chr9_+_73378057 | 0.06 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr18_+_30487264 | 0.06 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr17_+_81922329 | 0.06 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr3_-_90751055 | 0.06 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr3_-_80000091 | 0.06 |
ENSRNOT00000079394
|
Madd
|
MAP-kinase activating death domain |
| chr5_+_155934490 | 0.06 |
ENSRNOT00000077933
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr17_-_22678253 | 0.06 |
ENSRNOT00000083122
|
LOC100362172
|
LRRGT00112-like |
| chr17_-_56068125 | 0.06 |
ENSRNOT00000022462
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr15_-_61648267 | 0.06 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr9_-_95108739 | 0.06 |
ENSRNOT00000070819
|
Usp40
|
ubiquitin specific peptidase 40 |
| chr3_+_80670140 | 0.06 |
ENSRNOT00000085614
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr16_+_71090045 | 0.06 |
ENSRNOT00000020832
|
Ddhd2
|
DDHD domain containing 2 |
| chr5_-_147412705 | 0.06 |
ENSRNOT00000010688
|
RGD1561149
|
similar to mKIAA1522 protein |
| chr14_+_42015347 | 0.06 |
ENSRNOT00000044017
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr8_-_6305033 | 0.06 |
ENSRNOT00000029887
|
Cep126
|
centrosomal protein 126 |
| chr2_+_58448917 | 0.06 |
ENSRNOT00000082562
|
Ranbp3l
|
RAN binding protein 3-like |
| chr13_-_83457888 | 0.06 |
ENSRNOT00000076289
ENSRNOT00000004065 |
Sft2d2
|
SFT2 domain containing 2 |
| chr5_-_147867583 | 0.06 |
ENSRNOT00000000139
|
Kpna6
|
karyopherin subunit alpha 6 |
| chr16_+_59564419 | 0.06 |
ENSRNOT00000083434
|
Lonrf1
|
LON peptidase N-terminal domain and ring finger 1 |
| chr9_+_84203072 | 0.06 |
ENSRNOT00000018882
|
Sgpp2
|
sphingosine-1-phosphate phosphatase 2 |
| chr6_+_106052212 | 0.06 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr7_+_23403891 | 0.06 |
ENSRNOT00000037918
|
Syn3
|
synapsin III |
| chr13_-_94859390 | 0.06 |
ENSRNOT00000005467
ENSRNOT00000079763 |
AABR07021840.1
|
|
| chr13_+_43818010 | 0.06 |
ENSRNOT00000005532
|
LOC100909664
|
centrosomal protein of 170 kDa-like |
| chr4_-_176606382 | 0.06 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr2_+_248398917 | 0.06 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr8_-_21481735 | 0.06 |
ENSRNOT00000038069
|
Zfp426
|
zinc finger protein 426 |
| chr13_-_88536728 | 0.05 |
ENSRNOT00000003950
|
Uhmk1
|
U2AF homology motif kinase 1 |
| chr2_-_257546799 | 0.05 |
ENSRNOT00000089370
ENSRNOT00000090367 |
Miga1
|
mitoguardin 1 |
| chr10_+_17327275 | 0.05 |
ENSRNOT00000005486
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr5_-_133959447 | 0.05 |
ENSRNOT00000011985
|
Cyp4x1
|
cytochrome P450, family 4, subfamily x, polypeptide 1 |
| chr2_+_231941794 | 0.05 |
ENSRNOT00000091475
|
AABR07013288.4
|
|
| chr3_+_56056925 | 0.05 |
ENSRNOT00000088351
ENSRNOT00000010508 |
Klhl23
|
kelch-like family member 23 |
| chr1_-_37886675 | 0.05 |
ENSRNOT00000024149
|
RGD1566325
|
similar to regulator of sex-limitation candidate 16 |
| chr11_-_43022565 | 0.05 |
ENSRNOT00000002285
|
Riox2
|
ribosomal oxygenase 2 |
| chr10_-_86645529 | 0.05 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr3_+_8547349 | 0.05 |
ENSRNOT00000079108
ENSRNOT00000091697 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr12_-_46493203 | 0.05 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr3_+_48082935 | 0.05 |
ENSRNOT00000087711
ENSRNOT00000067545 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr6_+_104291071 | 0.05 |
ENSRNOT00000006798
|
Slc39a9
|
solute carrier family 39, member 9 |
| chr19_+_38039564 | 0.05 |
ENSRNOT00000087491
|
Nfatc3
|
nuclear factor of activated T-cells 3 |
| chrX_+_9956370 | 0.05 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr19_-_37796089 | 0.05 |
ENSRNOT00000024891
|
Ranbp10
|
RAN binding protein 10 |
| chr2_-_178521038 | 0.05 |
ENSRNOT00000033107
|
Rxfp1
|
relaxin/insulin-like family peptide receptor 1 |
| chr5_-_25721072 | 0.05 |
ENSRNOT00000021839
|
Tmem67
|
transmembrane protein 67 |
| chr13_-_80775230 | 0.05 |
ENSRNOT00000091389
ENSRNOT00000004762 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr10_+_71536533 | 0.05 |
ENSRNOT00000088138
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr8_+_73682887 | 0.05 |
ENSRNOT00000057522
|
Vps13c
|
vacuolar protein sorting 13C |
| chr8_-_114010277 | 0.05 |
ENSRNOT00000045087
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr9_-_66019065 | 0.05 |
ENSRNOT00000088729
|
Als2
|
ALS2, alsin Rho guanine nucleotide exchange factor |
| chr2_+_86996798 | 0.05 |
ENSRNOT00000070821
|
Zfp455
|
zinc finger protein 455 |
| chr17_+_89452814 | 0.05 |
ENSRNOT00000058760
|
RGD1561231
|
similar to MAP/microtubule affinity-regulating kinase 4 (MAP/microtubule affinity-regulating kinase like 1) |
| chr1_-_53802658 | 0.05 |
ENSRNOT00000032667
|
Afdn
|
afadin, adherens junction formation factor |
| chrM_+_9870 | 0.05 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chrX_+_84064427 | 0.04 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr9_+_52693831 | 0.04 |
ENSRNOT00000050275
|
Wdr75
|
WD repeat domain 75 |
| chr20_+_3588497 | 0.04 |
ENSRNOT00000048103
|
Vars2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr8_-_48597867 | 0.04 |
ENSRNOT00000077958
|
Nlrx1
|
NLR family member X1 |
| chr14_+_108143869 | 0.04 |
ENSRNOT00000090003
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr11_+_20474483 | 0.04 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr18_-_43945273 | 0.04 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr8_-_47339343 | 0.04 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr2_-_96509424 | 0.04 |
ENSRNOT00000090866
|
Zc2hc1a
|
zinc finger, C2HC-type containing 1A |
| chr13_-_35668968 | 0.04 |
ENSRNOT00000042862
ENSRNOT00000003428 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr7_+_18668692 | 0.04 |
ENSRNOT00000009532
|
Kank3
|
KN motif and ankyrin repeat domains 3 |
| chr13_-_86671515 | 0.04 |
ENSRNOT00000082869
|
AABR07021704.1
|
|
| chr1_+_101603222 | 0.04 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr11_+_81757983 | 0.04 |
ENSRNOT00000088503
|
Tbccd1
|
TBCC domain containing 1 |
| chrX_-_76708878 | 0.04 |
ENSRNOT00000045534
|
Atrx
|
ATRX, chromatin remodeler |
| chr1_+_100297152 | 0.04 |
ENSRNOT00000026100
|
Shank1
|
SH3 and multiple ankyrin repeat domains 1 |
| chr3_+_48106099 | 0.04 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr12_-_52658275 | 0.04 |
ENSRNOT00000041981
|
Zfp605
|
zinc finger protein 605 |
| chr2_-_257038105 | 0.04 |
ENSRNOT00000071195
|
Ptgfr
|
prostaglandin F receptor |
| chr19_+_38039729 | 0.04 |
ENSRNOT00000089783
|
Nfatc3
|
nuclear factor of activated T-cells 3 |
| chrX_+_115563038 | 0.04 |
ENSRNOT00000087859
|
Alg13
|
ALG13, UDP-N-acetylglucosaminyltransferase subunit |
| chr8_-_8524643 | 0.04 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr13_-_98078946 | 0.04 |
ENSRNOT00000035875
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr18_+_30496318 | 0.04 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr1_+_224998172 | 0.04 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chrX_+_908044 | 0.04 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr3_-_22500765 | 0.04 |
ENSRNOT00000025443
|
Dennd1a
|
DENN domain containing 1A |
| chr2_+_30094421 | 0.04 |
ENSRNOT00000082226
|
AABR07007839.1
|
|
| chr14_-_34570356 | 0.04 |
ENSRNOT00000003021
ENSRNOT00000090595 |
Srd5a3
|
steroid 5 alpha-reductase 3 |
| chr1_+_64740487 | 0.04 |
ENSRNOT00000081213
|
LOC103691005
|
zinc finger protein 679-like |
| chr2_-_118882562 | 0.04 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr5_-_150163395 | 0.04 |
ENSRNOT00000079935
|
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr1_-_275876329 | 0.04 |
ENSRNOT00000047903
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr2_-_248789508 | 0.04 |
ENSRNOT00000090705
|
Pkn2
|
protein kinase N2 |
| chr1_-_275882444 | 0.04 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr7_-_142063212 | 0.04 |
ENSRNOT00000089912
|
Slc11a2
|
solute carrier family 11 member 2 |
| chr17_-_89881919 | 0.04 |
ENSRNOT00000090982
|
LOC100910957
|
acyl-CoA-binding domain-containing protein 5-like |
| chr1_-_175657485 | 0.04 |
ENSRNOT00000024561
|
rnf141
|
ring finger protein 141 |
| chr12_-_1195591 | 0.03 |
ENSRNOT00000001446
|
Stard13
|
StAR-related lipid transfer domain containing 13 |
| chrM_+_11736 | 0.03 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr17_-_87826421 | 0.03 |
ENSRNOT00000068156
|
Arhgap21
|
Rho GTPase activating protein 21 |
| chr17_-_69862110 | 0.03 |
ENSRNOT00000058312
|
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr3_+_148837814 | 0.03 |
ENSRNOT00000091808
|
Asxl1
|
additional sex combs like 1 |
| chr10_+_84966989 | 0.03 |
ENSRNOT00000013580
|
Scrn2
|
secernin 2 |
| chr14_-_3300200 | 0.03 |
ENSRNOT00000037931
|
Btbd8
|
BTB (POZ) domain containing 8 |
| chr20_+_3246739 | 0.03 |
ENSRNOT00000061299
|
RT1-T24-2
|
RT1 class I, locus T24, gene 2 |
| chr2_+_185846232 | 0.03 |
ENSRNOT00000023418
|
Lrba
|
LPS responsive beige-like anchor protein |
| chr17_-_76250537 | 0.03 |
ENSRNOT00000092693
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr1_+_72956026 | 0.03 |
ENSRNOT00000031462
|
Rdh13
|
retinol dehydrogenase 13 |
| chr13_+_87986240 | 0.03 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr20_-_13994794 | 0.03 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr10_-_36323055 | 0.03 |
ENSRNOT00000081026
|
Zfp354c
|
zinc finger protein 354C |
| chr14_-_28967980 | 0.03 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr8_-_116855062 | 0.03 |
ENSRNOT00000050344
|
Rnf123
|
ring finger protein 123 |
| chr9_-_45206427 | 0.03 |
ENSRNOT00000042227
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr14_-_45863877 | 0.03 |
ENSRNOT00000002977
|
Pgm2
|
phosphoglucomutase 2 |
| chr5_+_6373583 | 0.03 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr17_-_84247038 | 0.03 |
ENSRNOT00000068553
|
Nebl
|
nebulette |
| chr7_-_15072703 | 0.03 |
ENSRNOT00000087294
|
Zfp799
|
zinc finger protein 799 |
| chr9_+_60039297 | 0.03 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr7_+_41475163 | 0.03 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr6_+_108009251 | 0.03 |
ENSRNOT00000059073
|
Ptgr2
|
prostaglandin reductase 2 |
| chr8_+_79159370 | 0.03 |
ENSRNOT00000086841
|
Rfx7
|
regulatory factor X, 7 |
| chr1_+_94793195 | 0.03 |
ENSRNOT00000079174
|
AC120712.2
|
|
| chr18_+_86116044 | 0.03 |
ENSRNOT00000058160
ENSRNOT00000076159 |
Rttn
|
rotatin |
| chr11_+_15436269 | 0.03 |
ENSRNOT00000002150
ENSRNOT00000084928 |
Usp25
|
ubiquitin specific peptidase 25 |
| chr4_-_51943519 | 0.03 |
ENSRNOT00000060459
|
Pot1
|
protection of telomeres 1 |
| chr2_-_231648122 | 0.03 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr17_-_15467320 | 0.03 |
ENSRNOT00000072490
ENSRNOT00000093561 |
Ogn
|
osteoglycin |
| chr1_+_274688580 | 0.03 |
ENSRNOT00000020580
|
Shoc2
|
SHOC2 leucine-rich repeat scaffold protein |
| chr5_-_79570073 | 0.03 |
ENSRNOT00000011845
|
Tnfsf15
|
tumor necrosis factor superfamily member 15 |
| chr8_-_83280888 | 0.03 |
ENSRNOT00000052341
|
Gfral
|
GDNF family receptor alpha like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prrx1_Isx_Prrxl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.1 | 0.2 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.1 | 0.2 | GO:1904139 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.2 | GO:1900170 | negative regulation of glucocorticoid mediated signaling pathway(GO:1900170) |
| 0.0 | 0.3 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.0 | 0.2 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.2 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.1 | GO:0001915 | negative regulation of T cell mediated cytotoxicity(GO:0001915) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 0.2 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0060903 | positive regulation of meiosis I(GO:0060903) |
| 0.0 | 0.1 | GO:0038109 | Kit signaling pathway(GO:0038109) diapedesis(GO:0050904) |
| 0.0 | 0.1 | GO:0061091 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.2 | GO:0090336 | positive regulation of brown fat cell differentiation(GO:0090336) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.2 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.0 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0032483 | regulation of Rab protein signal transduction(GO:0032483) |
| 0.0 | 0.0 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.0 | GO:1901581 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 0.1 | GO:0034626 | fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.0 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.0 | 0.0 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.0 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.0 | GO:0015676 | nickel cation transport(GO:0015675) vanadium ion transport(GO:0015676) lead ion transport(GO:0015692) nickel cation transmembrane transport(GO:0035444) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0009644 | response to high light intensity(GO:0009644) |
| 0.0 | 0.0 | GO:1901750 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.0 | GO:0071691 | cardiac muscle thin filament assembly(GO:0071691) |
| 0.0 | 0.0 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0017196 | N-terminal peptidyl-methionine acetylation(GO:0017196) |
| 0.0 | 0.2 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.3 | GO:0042581 | specific granule(GO:0042581) |
| 0.0 | 0.1 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.0 | 0.3 | GO:0030126 | COPI vesicle coat(GO:0030126) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.0 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.0 | GO:0043541 | UDP-N-acetylglucosamine transferase complex(GO:0043541) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0070826 | paraferritin complex(GO:0070826) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0001642 | group III metabotropic glutamate receptor activity(GO:0001642) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0042392 | sphingosine-1-phosphate phosphatase activity(GO:0042392) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004723 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.0 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.0 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.0 | GO:0015100 | cadmium ion transmembrane transporter activity(GO:0015086) lead ion transmembrane transporter activity(GO:0015094) nickel cation transmembrane transporter activity(GO:0015099) vanadium ion transmembrane transporter activity(GO:0015100) ferrous iron uptake transmembrane transporter activity(GO:0015639) chromium ion transmembrane transporter activity(GO:0070835) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.1 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |