Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
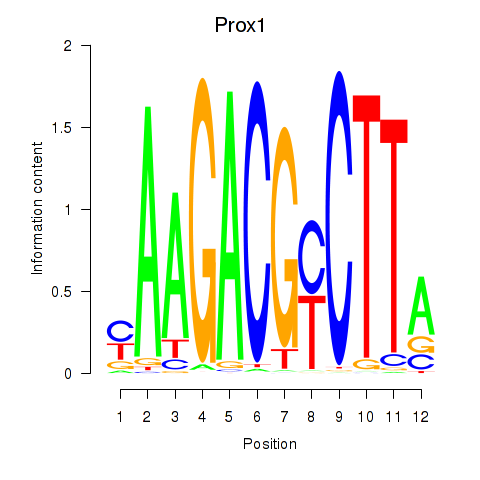
Results for Prox1
Z-value: 0.19
Transcription factors associated with Prox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prox1
|
ENSRNOG00000003694 | prospero homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prox1 | rn6_v1_chr13_+_108382399_108382399 | 0.64 | 2.4e-01 | Click! |
Activity profile of Prox1 motif
Sorted Z-values of Prox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_36865548 | 0.10 |
ENSRNOT00000076460
|
Parp4
|
poly (ADP-ribose) polymerase family, member 4 |
| chr2_-_140618405 | 0.06 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr1_+_212181374 | 0.06 |
ENSRNOT00000085921
|
Adgra1
|
adhesion G protein-coupled receptor A1 |
| chr12_+_13810180 | 0.06 |
ENSRNOT00000084865
ENSRNOT00000038203 |
Tnrc18
|
trinucleotide repeat containing 18 |
| chr13_+_51384562 | 0.05 |
ENSRNOT00000077928
|
Kdm5b
|
lysine demethylase 5B |
| chr11_+_66316606 | 0.05 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr15_+_47470863 | 0.05 |
ENSRNOT00000072438
|
LOC683422
|
similar to Plasma kallikrein precursor (Plasma prekallikrein) (Kininogenin) (Fletcher factor) |
| chr10_-_73787083 | 0.05 |
ENSRNOT00000035280
|
Med13
|
mediator complex subunit 13 |
| chr4_-_117126822 | 0.05 |
ENSRNOT00000086720
|
Rab11fip5
|
RAB11 family interacting protein 5 |
| chr7_-_114848414 | 0.04 |
ENSRNOT00000032620
|
Slc45a4
|
solute carrier family 45, member 4 |
| chr5_+_147069616 | 0.04 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr6_-_132608600 | 0.04 |
ENSRNOT00000015855
|
Degs2
|
delta(4)-desaturase, sphingolipid 2 |
| chr6_+_6619076 | 0.04 |
ENSRNOT00000042307
ENSRNOT00000080013 |
Eml4
|
echinoderm microtubule associated protein like 4 |
| chr17_-_76250537 | 0.04 |
ENSRNOT00000092693
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
| chr15_-_110046687 | 0.03 |
ENSRNOT00000057404
ENSRNOT00000006624 ENSRNOT00000089695 |
Nalcn
|
sodium leak channel, non-selective |
| chr8_-_5429581 | 0.03 |
ENSRNOT00000044174
|
Dync2h1
|
dynein cytoplasmic 2 heavy chain 1 |
| chr18_+_30880020 | 0.03 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr3_+_113131327 | 0.03 |
ENSRNOT00000018460
|
Tubgcp4
|
tubulin, gamma complex associated protein 4 |
| chr3_+_11593655 | 0.03 |
ENSRNOT00000074122
|
Pip5kl1
|
phosphatidylinositol-4-phosphate 5-kinase-like 1 |
| chr5_+_156613741 | 0.03 |
ENSRNOT00000076031
|
Sh2d5
|
SH2 domain containing 5 |
| chr5_+_151362019 | 0.03 |
ENSRNOT00000077083
ENSRNOT00000067801 |
Wasf2
|
WAS protein family, member 2 |
| chr5_+_47853818 | 0.03 |
ENSRNOT00000009228
ENSRNOT00000079656 |
Casp8ap2
|
caspase 8 associated protein 2 |
| chr3_-_162059524 | 0.03 |
ENSRNOT00000033873
|
Zfp334
|
zinc finger protein 334 |
| chr11_-_87769761 | 0.03 |
ENSRNOT00000052250
|
AABR07072266.1
|
|
| chr19_-_46101250 | 0.03 |
ENSRNOT00000015874
|
Adamts18
|
ADAM metallopeptidase with thrombospondin type 1 motif, 18 |
| chr4_+_25538497 | 0.02 |
ENSRNOT00000009264
|
Cfap69
|
cilia and flagella associated protein 69 |
| chr20_-_10257044 | 0.02 |
ENSRNOT00000068289
|
Wdr4
|
WD repeat domain 4 |
| chr16_+_32449116 | 0.02 |
ENSRNOT00000050549
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr14_+_91782354 | 0.02 |
ENSRNOT00000005902
|
Ikzf1
|
IKAROS family zinc finger 1 |
| chr3_-_133122691 | 0.02 |
ENSRNOT00000083637
|
Tasp1
|
taspase 1 |
| chr5_+_122019301 | 0.02 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr2_-_192697800 | 0.02 |
ENSRNOT00000066614
|
NEWGENE_2324572
|
small proline rich protein 4 |
| chr4_-_133951264 | 0.02 |
ENSRNOT00000090506
|
Pdzrn3
|
PDZ domain containing RING finger 3 |
| chr7_+_97760134 | 0.02 |
ENSRNOT00000086792
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr1_+_61374379 | 0.02 |
ENSRNOT00000015343
|
Zfp53
|
zinc finger protein 53 |
| chr6_-_42002819 | 0.02 |
ENSRNOT00000032417
|
Greb1
|
growth regulation by estrogen in breast cancer 1 |
| chr6_-_106971250 | 0.02 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr4_+_21872856 | 0.02 |
ENSRNOT00000044810
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chrX_+_156812064 | 0.02 |
ENSRNOT00000077142
|
Hcfc1
|
host cell factor C1 |
| chr3_-_110324084 | 0.02 |
ENSRNOT00000010381
|
Bmf
|
Bcl2 modifying factor |
| chr18_+_17091310 | 0.02 |
ENSRNOT00000093285
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr1_+_217459215 | 0.01 |
ENSRNOT00000092386
ENSRNOT00000092357 |
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr18_+_79773608 | 0.01 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr1_+_80920747 | 0.01 |
ENSRNOT00000026077
|
Zfp180
|
zinc finger protein 180 |
| chr19_-_49198073 | 0.01 |
ENSRNOT00000068440
|
Cdyl2
|
chromodomain Y-like 2 |
| chr8_-_118182559 | 0.01 |
ENSRNOT00000084838
|
Dhx30
|
DExH-box helicase 30 |
| chr16_+_19068783 | 0.01 |
ENSRNOT00000017424
|
Cherp
|
calcium homeostasis endoplasmic reticulum protein |
| chr1_+_243276403 | 0.01 |
ENSRNOT00000021496
|
Kank1
|
KN motif and ankyrin repeat domains 1 |
| chr10_+_3655038 | 0.01 |
ENSRNOT00000045047
|
Cpped1
|
calcineurin-like phosphoesterase domain containing 1 |
| chr6_-_10592454 | 0.01 |
ENSRNOT00000020600
|
Pigf
|
phosphatidylinositol glycan anchor biosynthesis, class F |
| chr1_+_199352344 | 0.01 |
ENSRNOT00000026466
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr15_-_28672530 | 0.01 |
ENSRNOT00000022593
ENSRNOT00000087722 |
Chd8
|
chromodomain helicase DNA binding protein 8 |
| chr10_-_57606171 | 0.01 |
ENSRNOT00000009015
|
Nup88
|
nucleoporin 88 |
| chr2_+_252018597 | 0.01 |
ENSRNOT00000020452
|
Mcoln2
|
mucolipin 2 |
| chr3_-_71295575 | 0.01 |
ENSRNOT00000040303
ENSRNOT00000083015 |
Zswim2
|
zinc finger, SWIM-type containing 2 |
| chr2_+_86996497 | 0.01 |
ENSRNOT00000042058
|
Zfp455
|
zinc finger protein 455 |
| chr4_+_61924013 | 0.01 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr2_-_77784522 | 0.01 |
ENSRNOT00000030328
|
LOC310177
|
similar to RIKEN cDNA 0610040D20 |
| chr7_-_126913585 | 0.01 |
ENSRNOT00000036025
|
Celsr1
|
cadherin, EGF LAG seven-pass G-type receptor 1 |
| chr4_+_89079014 | 0.01 |
ENSRNOT00000087451
|
Herc3
|
HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| chr13_+_51384389 | 0.00 |
ENSRNOT00000087025
|
Kdm5b
|
lysine demethylase 5B |
| chr1_+_265904566 | 0.00 |
ENSRNOT00000086041
ENSRNOT00000036156 |
Gbf1
|
golgi brefeldin A resistant guanine nucleotide exchange factor 1 |
| chr12_-_19328637 | 0.00 |
ENSRNOT00000001829
|
Taf6
|
TATA-box binding protein associated factor 6 |
| chr1_+_56982821 | 0.00 |
ENSRNOT00000059845
|
Ermard
|
ER membrane-associated RNA degradation |
| chr17_-_21705773 | 0.00 |
ENSRNOT00000078010
|
Gcnt2
|
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme |
| chr10_+_72147816 | 0.00 |
ENSRNOT00000090085
|
LOC102553386
|
SUMO-conjugating enzyme UBC9-like |
| chr10_+_81913689 | 0.00 |
ENSRNOT00000003780
|
Tob1
|
transducer of ErbB-2.1 |
| chr8_+_116776494 | 0.00 |
ENSRNOT00000050702
|
Cdhr4
|
cadherin-related family member 4 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0035937 | estrogen secretion(GO:0035937) estradiol secretion(GO:0035938) regulation of estrogen secretion(GO:2000861) regulation of estradiol secretion(GO:2000864) |
| 0.0 | 0.0 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0042284 | sphingolipid delta-4 desaturase activity(GO:0042284) |