Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
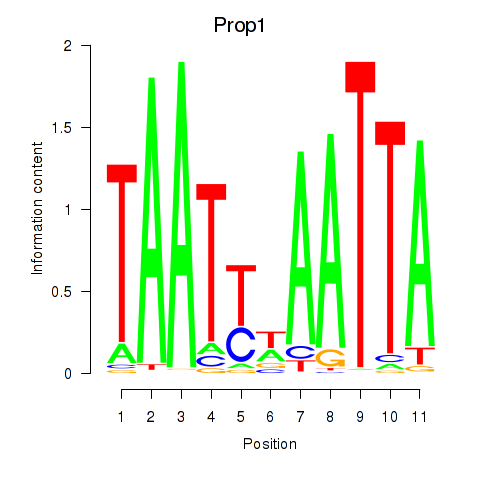
Results for Prop1
Z-value: 0.45
Transcription factors associated with Prop1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prop1
|
ENSRNOG00000003671 | PROP paired-like homeobox 1 |
Activity profile of Prop1 motif
Sorted Z-values of Prop1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_91054974 | 0.31 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr3_+_177397892 | 0.23 |
ENSRNOT00000078261
|
AABR07054983.1
|
|
| chrM_+_9870 | 0.21 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr4_+_51614676 | 0.16 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr2_-_149444548 | 0.15 |
ENSRNOT00000018600
|
P2ry12
|
purinergic receptor P2Y12 |
| chr5_+_6373583 | 0.14 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr2_+_148262110 | 0.14 |
ENSRNOT00000046641
|
AABR07010705.1
|
|
| chr1_-_163554839 | 0.14 |
ENSRNOT00000081509
|
Emsy
|
EMSY BRCA2-interacting transcriptional repressor |
| chr15_+_62406873 | 0.12 |
ENSRNOT00000047572
|
Olfm4
|
olfactomedin 4 |
| chr1_+_255040426 | 0.12 |
ENSRNOT00000092151
|
Pcgf5
|
polycomb group ring finger 5 |
| chr16_+_71090045 | 0.12 |
ENSRNOT00000020832
|
Ddhd2
|
DDHD domain containing 2 |
| chr1_+_64740487 | 0.11 |
ENSRNOT00000081213
|
LOC103691005
|
zinc finger protein 679-like |
| chr13_-_61591139 | 0.11 |
ENSRNOT00000005324
|
Rgs18
|
regulator of G-protein signaling 18 |
| chrX_+_118197217 | 0.10 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr3_+_155160481 | 0.09 |
ENSRNOT00000021133
|
Ppp1r16b
|
protein phosphatase 1, regulatory subunit 16B |
| chr1_-_167911961 | 0.09 |
ENSRNOT00000025097
|
Olr59
|
olfactory receptor 59 |
| chr1_-_219745654 | 0.09 |
ENSRNOT00000054848
|
LOC689065
|
hypothetical protein LOC689065 |
| chr10_+_31647898 | 0.09 |
ENSRNOT00000066536
|
LOC689968
|
similar to kidney injury molecule 1 |
| chr1_-_245817809 | 0.09 |
ENSRNOT00000080607
|
AABR07006672.1
|
|
| chr1_-_235405831 | 0.09 |
ENSRNOT00000071578
|
AABR07006458.1
|
|
| chr8_+_82038967 | 0.09 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr2_-_117454769 | 0.09 |
ENSRNOT00000068381
|
Mecom
|
MDS1 and EVI1 complex locus |
| chr1_-_66212418 | 0.09 |
ENSRNOT00000026074
|
LOC691722
|
hypothetical protein LOC691722 |
| chr9_-_7891514 | 0.09 |
ENSRNOT00000072684
|
Pot1b
|
protection of telomeres 1B |
| chr14_+_69800156 | 0.09 |
ENSRNOT00000072746
|
Lcorl
|
ligand dependent nuclear receptor corepressor-like |
| chr18_+_30487264 | 0.08 |
ENSRNOT00000040125
|
Pcdhb10
|
protocadherin beta 10 |
| chr15_-_44442875 | 0.08 |
ENSRNOT00000017939
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chr2_-_231648122 | 0.08 |
ENSRNOT00000014962
|
Ank2
|
ankyrin 2 |
| chr1_+_99749936 | 0.08 |
ENSRNOT00000025299
|
Klk7
|
kallikrein-related peptidase 7 |
| chr14_+_39663421 | 0.08 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr6_+_99883959 | 0.08 |
ENSRNOT00000010588
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr4_-_22307453 | 0.07 |
ENSRNOT00000047126
|
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chr14_+_42714315 | 0.07 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr11_+_88095170 | 0.07 |
ENSRNOT00000041557
|
Ccdc116
|
coiled-coil domain containing 116 |
| chr3_+_8547349 | 0.07 |
ENSRNOT00000079108
ENSRNOT00000091697 |
Sptan1
|
spectrin, alpha, non-erythrocytic 1 |
| chr1_+_271229036 | 0.07 |
ENSRNOT00000064023
|
LOC103690044
|
glutathione S-transferase omega-2 |
| chr14_+_3204390 | 0.07 |
ENSRNOT00000088078
|
Glmn
|
glomulin, FKBP associated protein |
| chr17_-_69862110 | 0.07 |
ENSRNOT00000058312
|
Akr1cl
|
aldo-keto reductase family 1, member C-like |
| chr5_-_12172009 | 0.07 |
ENSRNOT00000061903
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr2_+_206314213 | 0.07 |
ENSRNOT00000056068
|
Bcl2l15
|
BCL2-like 15 |
| chr6_+_96834525 | 0.07 |
ENSRNOT00000077935
|
Hif1a
|
hypoxia inducible factor 1 alpha subunit |
| chrM_+_9451 | 0.07 |
ENSRNOT00000041241
|
Mt-nd3
|
mitochondrially encoded NADH dehydrogenase 3 |
| chr11_+_34865532 | 0.07 |
ENSRNOT00000050342
|
Dyrk1a
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr9_-_69878706 | 0.07 |
ENSRNOT00000035879
|
Ino80d
|
INO80 complex subunit D |
| chr9_+_73378057 | 0.07 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr5_+_118574801 | 0.07 |
ENSRNOT00000035949
|
Ube2u
|
ubiquitin-conjugating enzyme E2U (putative) |
| chr2_+_30094421 | 0.07 |
ENSRNOT00000082226
|
AABR07007839.1
|
|
| chr1_+_66959610 | 0.07 |
ENSRNOT00000072122
ENSRNOT00000045994 |
Vom1r48
|
vomeronasal 1 receptor 48 |
| chr7_+_27174882 | 0.07 |
ENSRNOT00000051181
|
Glt8d2
|
glycosyltransferase 8 domain containing 2 |
| chr6_+_135313008 | 0.07 |
ENSRNOT00000030864
|
Tecpr2
|
tectonin beta-propeller repeat containing 2 |
| chr15_+_56666012 | 0.07 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr1_-_53152866 | 0.06 |
ENSRNOT00000085501
|
Fgfr1op
|
Fgfr1 oncogene partner |
| chr7_+_130308532 | 0.06 |
ENSRNOT00000011941
|
Miox
|
myo-inositol oxygenase |
| chr11_-_70499200 | 0.06 |
ENSRNOT00000002439
|
Slc12a8
|
solute carrier family 12, member 8 |
| chr7_-_52404774 | 0.06 |
ENSRNOT00000082100
|
Nav3
|
neuron navigator 3 |
| chr1_-_62316450 | 0.06 |
ENSRNOT00000079171
|
AABR07001926.3
|
|
| chr8_-_102149912 | 0.06 |
ENSRNOT00000011263
|
RGD1309079
|
similar to Ab2-095 |
| chr13_-_82006005 | 0.06 |
ENSRNOT00000039581
|
Mettl11b
|
methyltransferase like 11B |
| chr16_+_71889235 | 0.06 |
ENSRNOT00000038266
|
Adam32
|
ADAM metallopeptidase domain 32 |
| chr2_-_27343998 | 0.06 |
ENSRNOT00000091418
|
Polk
|
DNA polymerase kappa |
| chr18_+_16146447 | 0.06 |
ENSRNOT00000022117
|
Galnt1
|
polypeptide N-acetylgalactosaminyltransferase 1 |
| chr14_-_66978499 | 0.06 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr10_-_21265026 | 0.06 |
ENSRNOT00000011922
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr17_-_56068125 | 0.06 |
ENSRNOT00000022462
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr3_-_138683318 | 0.06 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr7_-_107768072 | 0.06 |
ENSRNOT00000093189
|
Ndrg1
|
N-myc downstream regulated 1 |
| chr3_-_52510507 | 0.06 |
ENSRNOT00000091259
|
Scn1a
|
sodium voltage-gated channel alpha subunit 1 |
| chr4_-_150522351 | 0.06 |
ENSRNOT00000079053
|
Zfp9
|
zinc finger protein 9 |
| chr10_+_65552897 | 0.06 |
ENSRNOT00000056217
|
Spag5
|
sperm associated antigen 5 |
| chr1_+_253221812 | 0.06 |
ENSRNOT00000085880
ENSRNOT00000054753 |
Kif20b
|
kinesin family member 20B |
| chr9_+_53013413 | 0.06 |
ENSRNOT00000005313
|
Ankar
|
ankyrin and armadillo repeat containing |
| chr13_-_76049363 | 0.06 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr16_+_8737974 | 0.06 |
ENSRNOT00000064255
|
Ercc6
|
ERCC excision repair 6, chromatin remodeling factor |
| chr4_-_176606382 | 0.05 |
ENSRNOT00000065576
|
Recql
|
RecQ like helicase |
| chr1_+_27476375 | 0.05 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr19_-_23944820 | 0.05 |
ENSRNOT00000080337
|
Zfp330
|
zinc finger protein 330 |
| chr8_-_132753145 | 0.05 |
ENSRNOT00000007467
ENSRNOT00000008172 |
RGD1566368
|
similar to Solute carrier family 6 (neurotransmitter transporter), member 20 |
| chrX_+_14019961 | 0.05 |
ENSRNOT00000004785
|
Sytl5
|
synaptotagmin-like 5 |
| chr10_+_84966989 | 0.05 |
ENSRNOT00000013580
|
Scrn2
|
secernin 2 |
| chr17_+_81922329 | 0.05 |
ENSRNOT00000031542
|
Cacnb2
|
calcium voltage-gated channel auxiliary subunit beta 2 |
| chr10_+_109773890 | 0.05 |
ENSRNOT00000071823
|
AC131537.1
|
|
| chr2_+_123597340 | 0.05 |
ENSRNOT00000058552
|
RGD1307100
|
similar to RIKEN cDNA D630029K19 |
| chr10_-_35175647 | 0.05 |
ENSRNOT00000078960
|
Cnot6
|
CCR4-NOT transcription complex, subunit 6 |
| chr20_-_13994794 | 0.05 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr4_+_70252366 | 0.05 |
ENSRNOT00000073039
|
Chl1
|
cell adhesion molecule L1-like |
| chr3_-_431933 | 0.05 |
ENSRNOT00000033653
|
Spopl
|
speckle type BTB/POZ protein like |
| chr13_-_55561726 | 0.05 |
ENSRNOT00000000817
ENSRNOT00000090162 |
Nek7
|
NIMA-related kinase 7 |
| chr3_+_134440195 | 0.05 |
ENSRNOT00000072928
|
AABR07054000.1
|
|
| chr1_+_84758233 | 0.05 |
ENSRNOT00000072734
|
AABR07002775.1
|
|
| chr20_+_3588497 | 0.05 |
ENSRNOT00000048103
|
Vars2
|
valyl-tRNA synthetase 2, mitochondrial |
| chr2_+_166402838 | 0.05 |
ENSRNOT00000064244
|
Nmd3
|
NMD3 ribosome export adaptor |
| chr5_+_5616483 | 0.05 |
ENSRNOT00000011026
|
Ncoa2
|
nuclear receptor coactivator 2 |
| chr1_-_88162583 | 0.04 |
ENSRNOT00000087411
|
Catsperg
|
cation channel sperm associated auxiliary subunit gamma |
| chr2_-_38110567 | 0.04 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chr2_-_128675408 | 0.04 |
ENSRNOT00000019256
|
Sclt1
|
sodium channel and clathrin linker 1 |
| chr6_+_64252970 | 0.04 |
ENSRNOT00000093700
|
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr1_+_86972244 | 0.04 |
ENSRNOT00000036907
|
Rinl
|
Ras and Rab interactor-like |
| chr8_-_109621408 | 0.04 |
ENSRNOT00000087398
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chr7_+_58419197 | 0.04 |
ENSRNOT00000085829
|
Zfc3h1
|
zinc finger, C3H1-type containing |
| chr6_+_18880737 | 0.04 |
ENSRNOT00000003432
|
Alkbh8
|
alkB homolog 8, tRNA methyltransferase |
| chr8_-_78397123 | 0.04 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr19_-_37796089 | 0.04 |
ENSRNOT00000024891
|
Ranbp10
|
RAN binding protein 10 |
| chr16_+_32457521 | 0.04 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr15_-_61648267 | 0.04 |
ENSRNOT00000071805
|
Naa16
|
N(alpha)-acetyltransferase 16, NatA auxiliary subunit |
| chr9_+_60039297 | 0.04 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chr1_-_125209727 | 0.04 |
ENSRNOT00000031067
|
Fan1
|
FANCD2 and FANCI associated nuclease 1 |
| chr11_-_67702955 | 0.04 |
ENSRNOT00000084540
ENSRNOT00000078285 |
Kpna1
|
karyopherin subunit alpha 1 |
| chr1_-_255815733 | 0.04 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_+_4989295 | 0.04 |
ENSRNOT00000041541
|
Fam172a
|
family with sequence similarity 172, member A |
| chr5_-_9094616 | 0.04 |
ENSRNOT00000072210
|
Sgk3
|
serum/glucocorticoid regulated kinase family, member 3 |
| chr11_-_71136673 | 0.04 |
ENSRNOT00000042240
|
Fyttd1
|
forty-two-three domain containing 1 |
| chr3_-_124724252 | 0.04 |
ENSRNOT00000028885
|
Slc23a2
|
solute carrier family 23 member 2 |
| chrX_+_99852198 | 0.04 |
ENSRNOT00000082451
|
AABR07040412.1
|
|
| chrX_+_84064427 | 0.04 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr1_+_156552328 | 0.04 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr10_-_91661558 | 0.04 |
ENSRNOT00000043156
|
AABR07030521.1
|
|
| chr2_+_118910587 | 0.04 |
ENSRNOT00000065518
|
Zfp639
|
zinc finger protein 639 |
| chr4_+_79021872 | 0.04 |
ENSRNOT00000012677
ENSRNOT00000067125 |
Fam221a
|
family with sequence similarity 221, member A |
| chr17_-_53688966 | 0.04 |
ENSRNOT00000078643
|
LOC100912163
|
AT-rich interactive domain-containing protein 4B-like |
| chr17_+_70262363 | 0.04 |
ENSRNOT00000048933
|
Fam208b
|
family with sequence similarity 208, member B |
| chr7_+_78188912 | 0.04 |
ENSRNOT00000043079
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr4_-_55011415 | 0.04 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr6_-_51018050 | 0.04 |
ENSRNOT00000082691
|
Gpr22
|
G protein-coupled receptor 22 |
| chr1_-_242861767 | 0.04 |
ENSRNOT00000021185
|
Cbwd1
|
COBW domain containing 1 |
| chr5_-_83648044 | 0.04 |
ENSRNOT00000046725
|
RGD1561195
|
similar to ribosomal protein L31 |
| chr3_+_80670140 | 0.04 |
ENSRNOT00000085614
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr13_-_74520634 | 0.04 |
ENSRNOT00000077169
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr16_-_79700992 | 0.04 |
ENSRNOT00000016420
|
Kbtbd11
|
kelch repeat and BTB domain containing 11 |
| chr2_+_54660722 | 0.04 |
ENSRNOT00000060376
|
Mroh2b
|
maestro heat-like repeat family member 2B |
| chr3_-_134696654 | 0.04 |
ENSRNOT00000006454
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3 |
| chr1_-_170186462 | 0.03 |
ENSRNOT00000035798
|
Olr202
|
olfactory receptor 202 |
| chr6_-_77421286 | 0.03 |
ENSRNOT00000011453
|
Nkx2-1
|
NK2 homeobox 1 |
| chr18_-_43945273 | 0.03 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr1_+_154446156 | 0.03 |
ENSRNOT00000093052
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr9_+_62291405 | 0.03 |
ENSRNOT00000051463
|
Plcl1
|
phospholipase C-like 1 |
| chr17_+_53965562 | 0.03 |
ENSRNOT00000090387
|
Ggps1
|
geranylgeranyl diphosphate synthase 1 |
| chr6_-_86914883 | 0.03 |
ENSRNOT00000091202
|
Mis18bp1
|
MIS18 binding protein 1 |
| chr2_+_128675814 | 0.03 |
ENSRNOT00000058366
|
RGD1359508
|
similar to protein C33A12.3 |
| chr8_+_55504359 | 0.03 |
ENSRNOT00000059086
|
Btg4
|
BTG anti-proliferation factor 4 |
| chr2_+_92559929 | 0.03 |
ENSRNOT00000033404
|
tGap1
|
GTPase activating protein testicular GAP1 |
| chr2_-_248789508 | 0.03 |
ENSRNOT00000090705
|
Pkn2
|
protein kinase N2 |
| chr1_-_67302751 | 0.03 |
ENSRNOT00000041518
|
Vom1r42
|
vomeronasal 1 receptor 42 |
| chr20_+_25990304 | 0.03 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr11_-_29710849 | 0.03 |
ENSRNOT00000029345
|
Krtap11-1
|
keratin associated protein 11-1 |
| chr8_+_126975833 | 0.03 |
ENSRNOT00000088348
|
AABR07071701.1
|
|
| chr16_-_47537476 | 0.03 |
ENSRNOT00000050279
|
Cldn24
|
claudin 24 |
| chr14_-_60552990 | 0.03 |
ENSRNOT00000042757
|
Zcchc4
|
zinc finger CCHC-type containing 4 |
| chr19_-_11669578 | 0.03 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chrX_-_63291371 | 0.03 |
ENSRNOT00000082421
|
Eif2s3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr7_+_49385705 | 0.03 |
ENSRNOT00000006083
|
Lin7a
|
lin-7 homolog A, crumbs cell polarity complex component |
| chr5_+_154868534 | 0.03 |
ENSRNOT00000091442
|
Luzp1
|
leucine zipper protein 1 |
| chr10_+_62566732 | 0.03 |
ENSRNOT00000021368
|
Taok1
|
TAO kinase 1 |
| chr1_+_101474334 | 0.03 |
ENSRNOT00000064245
|
Tulp2
|
tubby-like protein 2 |
| chr4_+_176606649 | 0.03 |
ENSRNOT00000084700
|
Golt1b
|
golgi transport 1B |
| chr15_+_4209703 | 0.03 |
ENSRNOT00000082236
|
Ppp3cb
|
protein phosphatase 3 catalytic subunit beta |
| chr6_-_122239614 | 0.03 |
ENSRNOT00000005015
|
Galc
|
galactosylceramidase |
| chr7_-_68512397 | 0.03 |
ENSRNOT00000058036
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr8_-_8524643 | 0.03 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr13_-_98078946 | 0.03 |
ENSRNOT00000035875
|
Ahctf1
|
AT hook containing transcription factor 1 |
| chr1_-_264294630 | 0.03 |
ENSRNOT00000035758
|
Sec31b
|
SEC31 homolog B, COPII coat complex component |
| chr8_-_109560747 | 0.03 |
ENSRNOT00000087334
|
Ppp2r3a
|
protein phosphatase 2, regulatory subunit B'', alpha |
| chrX_-_37003642 | 0.03 |
ENSRNOT00000040770
ENSRNOT00000058834 ENSRNOT00000058833 |
Adgrg2
|
adhesion G protein-coupled receptor G2 |
| chr1_-_53802658 | 0.03 |
ENSRNOT00000032667
|
Afdn
|
afadin, adherens junction formation factor |
| chr8_-_90984224 | 0.03 |
ENSRNOT00000044931
|
Lca5
|
LCA5, lebercilin |
| chr11_+_74834050 | 0.03 |
ENSRNOT00000002333
|
Atp13a4
|
ATPase 13A4 |
| chr6_+_8284878 | 0.03 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr1_-_217631862 | 0.03 |
ENSRNOT00000078514
ENSRNOT00000079805 |
Cttn
|
cortactin |
| chr8_+_49354115 | 0.02 |
ENSRNOT00000032837
|
Mpzl3
|
myelin protein zero-like 3 |
| chr2_-_186232292 | 0.02 |
ENSRNOT00000087088
|
Dclk2
|
doublecortin-like kinase 2 |
| chrX_-_152367861 | 0.02 |
ENSRNOT00000089414
|
LOC103690371
|
melanoma-associated antigen 10-like |
| chr13_-_82005741 | 0.02 |
ENSRNOT00000076404
|
Mettl11b
|
methyltransferase like 11B |
| chrX_+_124894466 | 0.02 |
ENSRNOT00000080894
|
Mcts1
|
MCTS1, re-initiation and release factor |
| chr3_+_47677720 | 0.02 |
ENSRNOT00000065340
|
Tbr1
|
T-box, brain, 1 |
| chr1_-_67134827 | 0.02 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr7_-_114590119 | 0.02 |
ENSRNOT00000079599
|
Ptk2
|
protein tyrosine kinase 2 |
| chr3_-_105663457 | 0.02 |
ENSRNOT00000067646
|
Zfp770
|
zinc finger protein 770 |
| chrX_+_9956370 | 0.02 |
ENSRNOT00000082316
|
LOC100910506
|
peripheral plasma membrane protein CASK-like |
| chr8_+_21905865 | 0.02 |
ENSRNOT00000027973
|
Ppan
|
peter pan homolog (Drosophila) |
| chr14_-_45863877 | 0.02 |
ENSRNOT00000002977
|
Pgm2
|
phosphoglucomutase 2 |
| chr12_-_46493203 | 0.02 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr18_+_40596166 | 0.02 |
ENSRNOT00000087984
|
Eif1a
|
eukaryotic translation initiation factor 1A |
| chr8_+_114300257 | 0.02 |
ENSRNOT00000090707
ENSRNOT00000018654 |
Pik3r4
|
phosphoinositide-3-kinase, regulatory subunit 4 |
| chr2_+_22910236 | 0.02 |
ENSRNOT00000078266
|
Homer1
|
homer scaffolding protein 1 |
| chr9_+_66952720 | 0.02 |
ENSRNOT00000023678
|
Nbeal1
|
neurobeachin-like 1 |
| chr9_+_52693831 | 0.02 |
ENSRNOT00000050275
|
Wdr75
|
WD repeat domain 75 |
| chr17_+_90802393 | 0.02 |
ENSRNOT00000003535
|
Edaradd
|
EDAR-associated death domain |
| chr4_-_61720956 | 0.02 |
ENSRNOT00000012879
|
Akr1b1
|
aldo-keto reductase family 1 member B |
| chr17_-_46794845 | 0.02 |
ENSRNOT00000077910
ENSRNOT00000090663 ENSRNOT00000078331 |
Elmo1
|
engulfment and cell motility 1 |
| chr2_+_34546988 | 0.02 |
ENSRNOT00000072577
|
Adamts6
|
ADAM metallopeptidase with thrombospondin type 1 motif, 6 |
| chr16_-_22853022 | 0.02 |
ENSRNOT00000050775
|
Ccdc94
|
coiled-coil domain containing 94 |
| chr2_+_198303168 | 0.02 |
ENSRNOT00000056262
|
Mtmr11
|
myotubularin related protein 11 |
| chr14_-_44078897 | 0.02 |
ENSRNOT00000031792
|
N4bp2
|
NEDD4 binding protein 2 |
| chr8_+_84945444 | 0.02 |
ENSRNOT00000008244
|
Klhl31
|
kelch-like family member 31 |
| chr13_-_92988137 | 0.02 |
ENSRNOT00000004803
|
Grem2
|
gremlin 2, DAN family BMP antagonist |
| chr11_+_71151132 | 0.02 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr3_+_81134505 | 0.02 |
ENSRNOT00000067664
|
Phf21a
|
PHD finger protein 21A |
| chr6_-_50923510 | 0.02 |
ENSRNOT00000010631
|
Bcap29
|
B-cell receptor-associated protein 29 |
| chr7_+_91384187 | 0.02 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
| chr13_-_50746279 | 0.02 |
ENSRNOT00000064556
ENSRNOT00000076856 ENSRNOT00000076673 |
Optc
|
opticin |
| chr3_+_154910626 | 0.02 |
ENSRNOT00000079515
|
Ralgapb
|
Ral GTPase activating protein non-catalytic beta subunit |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prop1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.2 | GO:0007208 | phospholipase C-activating serotonin receptor signaling pathway(GO:0007208) |
| 0.1 | 0.2 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.1 | GO:1902309 | negative regulation of peptidyl-serine dephosphorylation(GO:1902309) |
| 0.0 | 0.1 | GO:1900135 | positive regulation of renin secretion into blood stream(GO:1900135) |
| 0.0 | 0.1 | GO:0072695 | negative regulation of DNA recombination at telomere(GO:0048239) regulation of DNA recombination at telomere(GO:0072695) |
| 0.0 | 0.1 | GO:0006480 | N-terminal protein amino acid methylation(GO:0006480) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0048880 | medulla oblongata development(GO:0021550) sensory system development(GO:0048880) |
| 0.0 | 0.1 | GO:0071245 | cellular response to carbon monoxide(GO:0071245) cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
| 0.0 | 0.1 | GO:0090247 | cell motility involved in somitogenic axis elongation(GO:0090247) regulation of cell motility involved in somitogenic axis elongation(GO:0090249) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.1 | GO:0071623 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.1 | GO:0002803 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:1902412 | regulation of mitotic cytokinesis(GO:1902412) |
| 0.0 | 0.1 | GO:0043215 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:1901748 | leukotriene D4 metabolic process(GO:1901748) leukotriene D4 biosynthetic process(GO:1901750) |
| 0.0 | 0.1 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.0 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.1 | GO:0006362 | transcription elongation from RNA polymerase I promoter(GO:0006362) |
| 0.0 | 0.0 | GO:0015882 | L-ascorbic acid transport(GO:0015882) transepithelial L-ascorbic acid transport(GO:0070904) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) protein O-linked glycosylation via threonine(GO:0018243) |
| 0.0 | 0.0 | GO:1902962 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902962) negative regulation of metalloendopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902963) regulation of aspartic-type peptidase activity(GO:1905245) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.0 | GO:0033385 | geranylgeranyl diphosphate metabolic process(GO:0033385) geranylgeranyl diphosphate biosynthetic process(GO:0033386) |
| 0.0 | 0.0 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.0 | GO:0097401 | synaptic vesicle lumen acidification(GO:0097401) |
| 0.0 | 0.0 | GO:0045900 | negative regulation of translational elongation(GO:0045900) |
| 0.0 | 0.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0042581 | specific granule(GO:0042581) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0071886 | 1-(4-iodo-2,5-dimethoxyphenyl)propan-2-amine binding(GO:0071886) |
| 0.0 | 0.2 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0071885 | N-terminal protein N-methyltransferase activity(GO:0071885) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.2 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) oxidoreductase activity, acting on phosphorus or arsenic in donors(GO:0030613) oxidoreductase activity, acting on phosphorus or arsenic in donors, disulfide as acceptor(GO:0030614) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.0 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.1 | GO:0004311 | farnesyltranstransferase activity(GO:0004311) |
| 0.0 | 0.0 | GO:0070890 | L-ascorbate:sodium symporter activity(GO:0008520) L-ascorbic acid transporter activity(GO:0015229) sodium-dependent L-ascorbate transmembrane transporter activity(GO:0070890) |
| 0.0 | 0.0 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.0 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |