Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
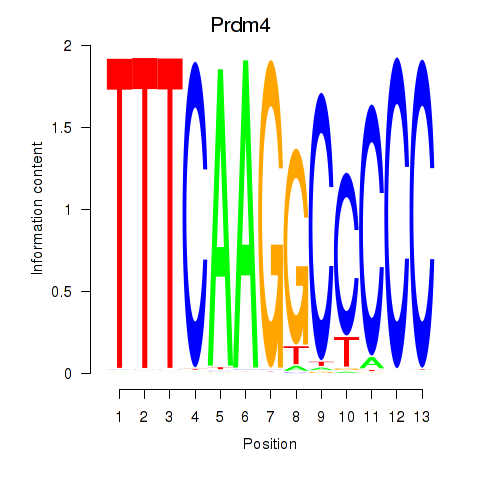
Results for Prdm4
Z-value: 0.11
Transcription factors associated with Prdm4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Prdm4
|
ENSRNOG00000004962 | PR/SET domain 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Prdm4 | rn6_v1_chr7_+_23960378_23960378 | -0.82 | 9.1e-02 | Click! |
Activity profile of Prdm4 motif
Sorted Z-values of Prdm4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_3012804 | 0.05 |
ENSRNOT00000061980
|
Arhgef33
|
Rho guanine nucleotide exchange factor 33 |
| chr4_+_26470864 | 0.04 |
ENSRNOT00000021979
|
Fzd1
|
frizzled class receptor 1 |
| chr1_-_89194602 | 0.03 |
ENSRNOT00000028518
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr1_-_89190128 | 0.03 |
ENSRNOT00000067813
|
Gapdhs
|
glyceraldehyde-3-phosphate dehydrogenase, spermatogenic |
| chr9_+_81672758 | 0.03 |
ENSRNOT00000020646
|
Ctdsp1
|
CTD small phosphatase 1 |
| chr3_+_11424099 | 0.02 |
ENSRNOT00000019184
|
Ptges2
|
prostaglandin E synthase 2 |
| chr19_-_37525762 | 0.02 |
ENSRNOT00000023606
|
Atp6v0d1
|
ATPase H+ transporting V0 subunit D1 |
| chr7_+_98432442 | 0.00 |
ENSRNOT00000038973
|
Fer1l6
|
fer-1-like family member 6 |
| chr1_-_107373807 | 0.00 |
ENSRNOT00000056024
|
Svip
|
small VCP interacting protein |
Network of associatons between targets according to the STRING database.
First level regulatory network of Prdm4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0035425 | autocrine signaling(GO:0035425) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004365 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |