Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
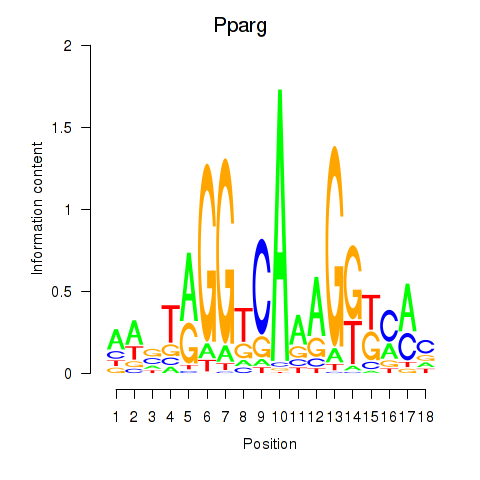
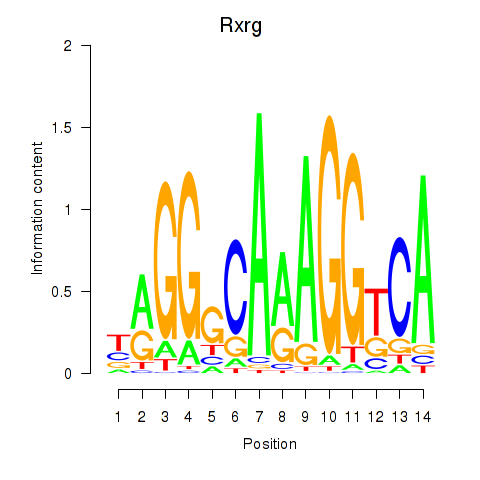
Results for Pparg_Rxrg
Z-value: 1.16
Transcription factors associated with Pparg_Rxrg
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pparg
|
ENSRNOG00000008839 | peroxisome proliferator-activated receptor gamma |
|
Rxrg
|
ENSRNOG00000004537 | retinoid X receptor gamma |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Rxrg | rn6_v1_chr13_+_85818427_85818473 | -0.61 | 2.8e-01 | Click! |
| Pparg | rn6_v1_chr4_+_147333056_147333056 | 0.39 | 5.2e-01 | Click! |
Activity profile of Pparg_Rxrg motif
Sorted Z-values of Pparg_Rxrg motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_2174131 | 3.22 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr3_-_66417741 | 1.76 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr10_+_84167331 | 0.90 |
ENSRNOT00000010965
|
Hoxb4
|
homeo box B4 |
| chr1_-_226049929 | 0.88 |
ENSRNOT00000007320
|
Best1
|
bestrophin 1 |
| chr10_-_15603649 | 0.75 |
ENSRNOT00000051483
|
Hba-a2
|
hemoglobin alpha, adult chain 2 |
| chr10_+_59533480 | 0.69 |
ENSRNOT00000087723
|
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr7_-_119441487 | 0.62 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr5_-_172809353 | 0.61 |
ENSRNOT00000022246
|
Gabrd
|
gamma-aminobutyric acid type A receptor delta subunit |
| chr19_+_41482728 | 0.58 |
ENSRNOT00000022943
|
Calb2
|
calbindin 2 |
| chr3_+_123106694 | 0.56 |
ENSRNOT00000028829
|
Oxt
|
oxytocin/neurophysin 1 prepropeptide |
| chr3_+_149624712 | 0.53 |
ENSRNOT00000018581
|
Bpifa1
|
BPI fold containing family A, member 1 |
| chr7_-_143793774 | 0.52 |
ENSRNOT00000079678
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr1_-_166939541 | 0.50 |
ENSRNOT00000079675
|
Folr1
|
folate receptor 1 |
| chr12_-_18526250 | 0.49 |
ENSRNOT00000001791
|
Asmt
|
acetylserotonin O-methyltransferase |
| chr10_+_56662561 | 0.49 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr6_+_43001948 | 0.48 |
ENSRNOT00000007374
|
Hpcal1
|
hippocalcin-like 1 |
| chr10_+_56662242 | 0.47 |
ENSRNOT00000086919
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr7_-_143793970 | 0.46 |
ENSRNOT00000016205
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr3_-_7498555 | 0.44 |
ENSRNOT00000017725
|
Barhl1
|
BarH-like homeobox 1 |
| chr1_-_262013619 | 0.43 |
ENSRNOT00000021278
|
Hps1
|
HPS1, biogenesis of lysosomal organelles complex 3 subunit 1 |
| chrX_-_40086870 | 0.43 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr6_+_123361864 | 0.42 |
ENSRNOT00000057577
|
AABR07065353.1
|
|
| chr5_-_152473868 | 0.41 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr3_+_161121697 | 0.41 |
ENSRNOT00000036020
|
Wfdc10a
|
WAP four-disulfide core domain 10A |
| chr7_+_144647587 | 0.40 |
ENSRNOT00000022398
|
Hoxc4
|
homeo box C4 |
| chr10_+_103396155 | 0.40 |
ENSRNOT00000086924
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr16_-_20890949 | 0.39 |
ENSRNOT00000081977
|
Homer3
|
homer scaffolding protein 3 |
| chr2_-_225107283 | 0.39 |
ENSRNOT00000055711
|
Slc44a3
|
solute carrier family 44, member 3 |
| chr7_+_120153184 | 0.39 |
ENSRNOT00000013538
|
Lgals1
|
galectin 1 |
| chr7_+_117951154 | 0.39 |
ENSRNOT00000060181
|
Znf7
|
zinc finger protein 7 |
| chr9_-_65957101 | 0.36 |
ENSRNOT00000014094
|
Mpp4
|
membrane palmitoylated protein 4 |
| chr10_+_86399827 | 0.35 |
ENSRNOT00000009299
|
Grb7
|
growth factor receptor bound protein 7 |
| chr19_-_37250919 | 0.34 |
ENSRNOT00000021015
|
Exoc3l1
|
exocyst complex component 3-like 1 |
| chr10_+_13230158 | 0.33 |
ENSRNOT00000075648
|
Csap1
|
common salivary protein 1 |
| chr12_-_48627297 | 0.32 |
ENSRNOT00000000890
|
Iscu
|
iron-sulfur cluster assembly enzyme |
| chr5_-_156734541 | 0.32 |
ENSRNOT00000021036
|
LOC100909857
|
cytidine deaminase-like |
| chr12_+_49565992 | 0.31 |
ENSRNOT00000071328
|
Crybb3
|
crystallin, beta B3 |
| chr8_-_22974321 | 0.31 |
ENSRNOT00000017369
|
Epor
|
erythropoietin receptor |
| chr10_-_15311189 | 0.31 |
ENSRNOT00000027371
|
Prr35
|
proline rich 35 |
| chr20_-_3819200 | 0.31 |
ENSRNOT00000000542
ENSRNOT00000081486 |
Hsd17b8
|
hydroxysteroid (17-beta) dehydrogenase 8 |
| chr20_+_13827660 | 0.30 |
ENSRNOT00000093131
|
Ddt
|
D-dopachrome tautomerase |
| chr3_-_1584946 | 0.29 |
ENSRNOT00000031058
|
Pax8
|
paired box 8 |
| chr1_-_89488223 | 0.29 |
ENSRNOT00000028624
|
Fxyd1
|
FXYD domain-containing ion transport regulator 1 |
| chr4_+_100407658 | 0.29 |
ENSRNOT00000018562
|
Capg
|
capping actin protein, gelsolin like |
| chrX_-_123662350 | 0.29 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr10_+_103395511 | 0.29 |
ENSRNOT00000004256
|
Gprc5c
|
G protein-coupled receptor, class C, group 5, member C |
| chr14_+_83752393 | 0.29 |
ENSRNOT00000081123
|
Selenom
|
selenoprotein M |
| chr1_-_190965115 | 0.29 |
ENSRNOT00000023483
|
AC103221.1
|
|
| chr3_+_153197644 | 0.29 |
ENSRNOT00000008386
|
Tldc2
|
TBC/LysM-associated domain containing 2 |
| chr3_+_172195844 | 0.28 |
ENSRNOT00000034915
|
Npepl1
|
aminopeptidase-like 1 |
| chr9_+_14529218 | 0.28 |
ENSRNOT00000016893
|
Apobec2
|
apolipoprotein B mRNA editing enzyme catalytic subunit 2 |
| chr3_+_159887036 | 0.28 |
ENSRNOT00000031868
|
R3hdml
|
R3H domain containing-like |
| chr13_-_102721218 | 0.28 |
ENSRNOT00000005459
|
Marc1
|
mitochondrial amidoxime reducing component 1 |
| chr5_+_137257287 | 0.27 |
ENSRNOT00000037160
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr10_+_104437648 | 0.26 |
ENSRNOT00000035001
|
AC130970.1
|
|
| chr20_+_13662867 | 0.26 |
ENSRNOT00000032257
|
RGD1564162
|
similar to Homo sapiens fetal lung specific expression unknown |
| chr3_+_154786215 | 0.25 |
ENSRNOT00000019787
|
Lbp
|
lipopolysaccharide binding protein |
| chr12_-_21746236 | 0.25 |
ENSRNOT00000001869
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr8_-_22785671 | 0.25 |
ENSRNOT00000045384
|
Spc24
|
SPC24, NDC80 kinetochore complex component |
| chr18_+_63130542 | 0.25 |
ENSRNOT00000024947
|
Tubb6
|
tubulin, beta 6 class V |
| chr7_-_144960527 | 0.25 |
ENSRNOT00000086554
|
Zfp385a
|
zinc finger protein 385A |
| chr7_-_70842405 | 0.24 |
ENSRNOT00000047449
|
Nxph4
|
neurexophilin 4 |
| chr2_-_187401786 | 0.24 |
ENSRNOT00000025624
|
Hapln2
|
hyaluronan and proteoglycan link protein 2 |
| chr8_+_70112925 | 0.24 |
ENSRNOT00000082401
|
Megf11
|
multiple EGF-like-domains 11 |
| chr8_-_60086403 | 0.24 |
ENSRNOT00000020544
|
Etfa
|
electron transfer flavoprotein alpha subunit |
| chr2_-_187863503 | 0.24 |
ENSRNOT00000093036
ENSRNOT00000026705 ENSRNOT00000082174 |
Lmna
|
lamin A/C |
| chr5_-_148392689 | 0.23 |
ENSRNOT00000018464
ENSRNOT00000080166 |
Tinagl1
|
tubulointerstitial nephritis antigen-like 1 |
| chr1_+_71434819 | 0.23 |
ENSRNOT00000059069
|
Zfp787
|
zinc finger protein 787 |
| chr6_+_26051396 | 0.23 |
ENSRNOT00000006452
|
Rbks
|
ribokinase |
| chr5_+_167288223 | 0.23 |
ENSRNOT00000081579
|
Eno1
|
enolase 1 |
| chrX_-_10218583 | 0.22 |
ENSRNOT00000013382
|
Nyx
|
nyctalopin |
| chr8_-_68312909 | 0.22 |
ENSRNOT00000066106
|
RGD1309779
|
similar to ENSANGP00000021391 |
| chr2_+_61991742 | 0.21 |
ENSRNOT00000046715
|
LOC499544
|
LRRGT00154 |
| chr9_+_80118029 | 0.21 |
ENSRNOT00000023068
|
Igfbp2
|
insulin-like growth factor binding protein 2 |
| chr14_+_46001849 | 0.21 |
ENSRNOT00000076611
|
Rell1
|
RELT-like 1 |
| chr10_+_71202456 | 0.21 |
ENSRNOT00000076893
|
Hnf1b
|
HNF1 homeobox B |
| chr2_-_210116038 | 0.21 |
ENSRNOT00000074950
|
LOC684509
|
similar to NADH-ubiquinone oxidoreductase B9 subunit (Complex I-B9) (CI-B9) |
| chr13_-_50535389 | 0.20 |
ENSRNOT00000076506
|
Kiss1
|
KiSS-1 metastasis-suppressor |
| chr7_+_3173287 | 0.20 |
ENSRNOT00000092037
|
Pym1
|
PYM homolog 1, exon junction complex associated factor |
| chr3_+_8832740 | 0.20 |
ENSRNOT00000022552
|
Phyhd1
|
phytanoyl-CoA dioxygenase domain containing 1 |
| chr8_+_98755104 | 0.20 |
ENSRNOT00000056562
|
Zic4
|
Zic family member 4 |
| chr15_+_24159647 | 0.20 |
ENSRNOT00000082675
|
Lgals3
|
galectin 3 |
| chr9_-_82477136 | 0.20 |
ENSRNOT00000026753
ENSRNOT00000089555 |
Resp18
|
regulated endocrine-specific protein 18 |
| chr3_+_81287242 | 0.19 |
ENSRNOT00000086530
|
Pex16
|
peroxisomal biogenesis factor 16 |
| chr1_-_89369960 | 0.19 |
ENSRNOT00000028545
|
Hamp
|
hepcidin antimicrobial peptide |
| chr3_-_46361092 | 0.19 |
ENSRNOT00000008987
|
Cd302
|
CD302 molecule |
| chr14_-_18733391 | 0.19 |
ENSRNOT00000003745
|
Cxcl2
|
C-X-C motif chemokine ligand 2 |
| chr1_-_187779675 | 0.19 |
ENSRNOT00000024648
|
Arl6ip1
|
ADP-ribosylation factor like GTPase 6 interacting protein 1 |
| chr3_+_1452644 | 0.19 |
ENSRNOT00000007949
|
Il1rn
|
interleukin 1 receptor antagonist |
| chr9_+_69953440 | 0.19 |
ENSRNOT00000034740
|
Eef1b2
|
eukaryotic translation elongation factor 1 beta 2 |
| chr2_-_154334356 | 0.19 |
ENSRNOT00000057724
|
Plch1
|
phospholipase C, eta 1 |
| chr5_+_154522119 | 0.19 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr1_-_89543967 | 0.18 |
ENSRNOT00000079631
|
Hpn
|
hepsin |
| chr10_-_44746549 | 0.18 |
ENSRNOT00000003841
|
Fam183b
|
family with sequence similarity 183, member B |
| chr10_+_94566928 | 0.18 |
ENSRNOT00000078446
|
Prr29
|
proline rich 29 |
| chr10_+_23661343 | 0.18 |
ENSRNOT00000047970
|
Ebf1
|
early B-cell factor 1 |
| chr1_+_219833299 | 0.18 |
ENSRNOT00000087432
|
Pc
|
pyruvate carboxylase |
| chr13_-_89745835 | 0.18 |
ENSRNOT00000005352
|
Klhdc9
|
kelch domain containing 9 |
| chr17_+_42302540 | 0.18 |
ENSRNOT00000025389
|
Gmnn
|
geminin, DNA replication inhibitor |
| chr9_-_65951516 | 0.18 |
ENSRNOT00000038132
|
Mpp4
|
membrane palmitoylated protein 4 |
| chr10_+_11146359 | 0.18 |
ENSRNOT00000006314
|
Pam16
|
presequence translocase associated motor 16 homolog |
| chr5_+_129257429 | 0.18 |
ENSRNOT00000072396
|
Ttc39a
|
tetratricopeptide repeat domain 39A |
| chr5_-_154438361 | 0.17 |
ENSRNOT00000085003
|
AC141344.2
|
|
| chr13_-_89306219 | 0.17 |
ENSRNOT00000004183
ENSRNOT00000079247 |
Fcrla
|
Fc receptor-like A |
| chr11_+_87204175 | 0.17 |
ENSRNOT00000000306
|
Slc25a1
|
solute carrier family 25 member 1 |
| chr11_+_65022100 | 0.17 |
ENSRNOT00000003934
|
Nr1i2
|
nuclear receptor subfamily 1, group I, member 2 |
| chr15_+_38341089 | 0.17 |
ENSRNOT00000015367
|
Fgf9
|
fibroblast growth factor 9 |
| chr7_-_120490061 | 0.17 |
ENSRNOT00000016247
|
Slc16a8
|
solute carrier family 16 member 8 |
| chr10_-_19652898 | 0.17 |
ENSRNOT00000009648
|
Spdl1
|
spindle apparatus coiled-coil protein 1 |
| chr19_+_37229120 | 0.17 |
ENSRNOT00000076335
|
Hsf4
|
heat shock transcription factor 4 |
| chr18_+_3565166 | 0.16 |
ENSRNOT00000039377
|
Riok3
|
RIO kinase 3 |
| chr1_+_168964202 | 0.16 |
ENSRNOT00000089102
|
LOC103694855
|
hemoglobin subunit beta-2-like |
| chr9_+_73493027 | 0.16 |
ENSRNOT00000074427
ENSRNOT00000089478 |
Unc80
|
unc-80 homolog, NALCN activator |
| chr1_-_7480825 | 0.16 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr12_+_22274828 | 0.16 |
ENSRNOT00000001914
|
Epo
|
erythropoietin |
| chr8_-_108109801 | 0.16 |
ENSRNOT00000034277
|
Sox14
|
SRY box 14 |
| chr20_+_13778178 | 0.16 |
ENSRNOT00000058314
|
Gstt4
|
glutathione S-transferase, theta 4 |
| chr7_-_11400805 | 0.16 |
ENSRNOT00000027634
|
Dapk3
|
death-associated protein kinase 3 |
| chr6_-_26051229 | 0.16 |
ENSRNOT00000005855
|
Bre
|
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chr1_+_163445527 | 0.16 |
ENSRNOT00000020520
|
Lrrc32
|
leucine rich repeat containing 32 |
| chr7_+_119626637 | 0.16 |
ENSRNOT00000000201
|
Mpst
|
mercaptopyruvate sulfurtransferase |
| chr18_-_29340403 | 0.16 |
ENSRNOT00000025157
|
Hbegf
|
heparin-binding EGF-like growth factor |
| chr1_+_88955440 | 0.16 |
ENSRNOT00000091101
|
Prodh2
|
proline dehydrogenase 2 |
| chr3_-_153454160 | 0.16 |
ENSRNOT00000010298
|
Ghrh
|
growth hormone releasing hormone |
| chr10_-_59743315 | 0.16 |
ENSRNOT00000093646
|
Emc6
|
ER membrane protein complex subunit 6 |
| chr3_+_14482388 | 0.15 |
ENSRNOT00000025857
|
Gsn
|
gelsolin |
| chr1_+_78659435 | 0.15 |
ENSRNOT00000071271
|
Ceacam9
|
carcinoembryonic antigen-related cell adhesion molecule 9 |
| chr1_-_24191908 | 0.15 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr13_-_44345735 | 0.15 |
ENSRNOT00000005006
|
Tmem163
|
transmembrane protein 163 |
| chr7_-_34485034 | 0.15 |
ENSRNOT00000007351
|
Snrpf
|
small nuclear ribonucleoprotein polypeptide F |
| chr13_+_85580828 | 0.15 |
ENSRNOT00000005611
|
Aldh9a1
|
aldehyde dehydrogenase 9 family, member A1 |
| chr5_+_137243094 | 0.15 |
ENSRNOT00000067237
|
Med8
|
mediator complex subunit 8 |
| chr1_-_211956858 | 0.15 |
ENSRNOT00000054886
|
Cfap46
|
cilia and flagella associated protein 46 |
| chr3_-_2938883 | 0.15 |
ENSRNOT00000084196
|
LOC108348105
|
allergen Fel d 4-like |
| chr10_-_59743037 | 0.15 |
ENSRNOT00000026168
|
Emc6
|
ER membrane protein complex subunit 6 |
| chr5_+_70441123 | 0.14 |
ENSRNOT00000087517
|
Fsd1l
|
fibronectin type III and SPRY domain containing 1-like |
| chr11_-_84931160 | 0.14 |
ENSRNOT00000071848
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr8_+_72741155 | 0.14 |
ENSRNOT00000090977
|
Rps27l
|
ribosomal protein S27-like |
| chr12_-_30566032 | 0.14 |
ENSRNOT00000093378
|
Gbas
|
glioblastoma amplified sequence |
| chr1_+_213886775 | 0.14 |
ENSRNOT00000020864
|
Pkp3
|
plakophilin 3 |
| chr2_+_200452624 | 0.14 |
ENSRNOT00000026121
|
Hmgcs2
|
3-hydroxy-3-methylglutaryl-CoA synthase 2 |
| chr5_+_57947716 | 0.14 |
ENSRNOT00000067657
|
Dnai1
|
dynein, axonemal, intermediate chain 1 |
| chr9_+_64095978 | 0.14 |
ENSRNOT00000021533
|
Maip1
|
matrix AAA peptidase interacting protein 1 |
| chr10_-_103848035 | 0.14 |
ENSRNOT00000029001
|
Fads6
|
fatty acid desaturase 6 |
| chr7_-_130405347 | 0.14 |
ENSRNOT00000013985
|
Cpt1b
|
carnitine palmitoyltransferase 1B |
| chr7_-_116963281 | 0.14 |
ENSRNOT00000082058
ENSRNOT00000012306 |
Tsta3
|
tissue specific transplantation antigen P35B |
| chr8_+_48569328 | 0.14 |
ENSRNOT00000084030
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr7_-_117369159 | 0.13 |
ENSRNOT00000016424
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr9_+_70310932 | 0.13 |
ENSRNOT00000088268
|
LOC103690541
|
uncharacterized LOC103690541 |
| chr6_-_60151420 | 0.13 |
ENSRNOT00000073910
|
Ifrd1
|
interferon-related developmental regulator 1 |
| chr1_-_224986291 | 0.13 |
ENSRNOT00000026284
|
Taf6l
|
TATA-box binding protein associated factor 6 like |
| chr4_-_176679815 | 0.13 |
ENSRNOT00000090122
|
Gys2
|
glycogen synthase 2 |
| chr4_+_115046693 | 0.13 |
ENSRNOT00000031583
|
Bola3
|
bolA family member 3 |
| chr7_+_141889444 | 0.13 |
ENSRNOT00000079011
|
Atf1
|
activating transcription factor 1 |
| chr6_+_136358057 | 0.13 |
ENSRNOT00000092741
|
Klc1
|
kinesin light chain 1 |
| chr1_-_222178725 | 0.13 |
ENSRNOT00000028697
|
Esrra
|
estrogen related receptor, alpha |
| chr5_+_169364306 | 0.13 |
ENSRNOT00000014213
|
Acot7
|
acyl-CoA thioesterase 7 |
| chr14_+_11198896 | 0.13 |
ENSRNOT00000079767
|
Hnrnpdl
|
heterogeneous nuclear ribonucleoprotein D-like |
| chr10_+_105073077 | 0.13 |
ENSRNOT00000083022
ENSRNOT00000086382 ENSRNOT00000087084 |
Ten1
|
TEN1 CST complex subunit |
| chr16_+_81616604 | 0.13 |
ENSRNOT00000026392
ENSRNOT00000057740 |
Adprhl1
Grtp1
|
ADP-ribosylhydrolase like 1 growth hormone regulated TBC protein 1 |
| chr9_+_50526811 | 0.13 |
ENSRNOT00000036990
|
RGD1305645
|
similar to RIKEN cDNA 1500015O10 |
| chrX_+_136488691 | 0.13 |
ENSRNOT00000093432
|
Arhgap36
|
Rho GTPase activating protein 36 |
| chr1_+_72636959 | 0.13 |
ENSRNOT00000023489
|
Il11
|
interleukin 11 |
| chr8_+_99894762 | 0.13 |
ENSRNOT00000081768
|
Plscr4
|
phospholipid scramblase 4 |
| chr12_-_2661565 | 0.13 |
ENSRNOT00000064859
|
Cd209f
|
CD209f antigen |
| chr10_-_108196217 | 0.13 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr17_-_32558180 | 0.13 |
ENSRNOT00000022681
|
Serpinb1b
|
serine (or cysteine) peptidase inhibitor, clade B, member 1b |
| chr5_+_59008933 | 0.13 |
ENSRNOT00000023060
|
Car9
|
carbonic anhydrase 9 |
| chr3_-_165360292 | 0.13 |
ENSRNOT00000065615
|
Nfatc2
|
nuclear factor of activated T-cells 2 |
| chr5_+_137257637 | 0.13 |
ENSRNOT00000093001
|
Elovl1
|
ELOVL fatty acid elongase 1 |
| chr19_+_24800072 | 0.13 |
ENSRNOT00000005470
|
Ptger1
|
prostaglandin E receptor 1 |
| chr6_+_107460668 | 0.12 |
ENSRNOT00000013515
|
Acot2
|
acyl-CoA thioesterase 2 |
| chr10_-_45923829 | 0.12 |
ENSRNOT00000035285
|
Olr1462
|
olfactory receptor 1462 |
| chr3_+_176691329 | 0.12 |
ENSRNOT00000017319
|
Ppdpf
|
pancreatic progenitor cell differentiation and proliferation factor |
| chr1_-_91042230 | 0.12 |
ENSRNOT00000073107
|
LOC103690054
|
DNA-directed RNA polymerase II subunit RPB9 |
| chrX_-_113584459 | 0.12 |
ENSRNOT00000025923
|
Kcne5
|
potassium voltage-gated channel subfamily E regulatory subunit 5 |
| chr4_-_145390447 | 0.12 |
ENSRNOT00000091965
ENSRNOT00000012136 |
Cidec
|
cell death-inducing DFFA-like effector c |
| chr20_+_46667454 | 0.12 |
ENSRNOT00000065890
|
Sesn1
|
sestrin 1 |
| chr9_-_50820290 | 0.12 |
ENSRNOT00000050748
|
LOC102548013
|
uncharacterized LOC102548013 |
| chr1_-_89360733 | 0.12 |
ENSRNOT00000028544
|
Mag
|
myelin-associated glycoprotein |
| chr4_-_181477281 | 0.12 |
ENSRNOT00000055463
|
Mansc4
|
MANSC domain containing 4 |
| chr6_-_33738825 | 0.12 |
ENSRNOT00000039492
|
Slc7a15
|
solute carrier family 7 (cationic amino acid transporter, y+ system), member 15 |
| chr6_+_76174452 | 0.12 |
ENSRNOT00000084062
|
Psma6
|
proteasome subunit alpha 6 |
| chr2_-_148722263 | 0.12 |
ENSRNOT00000017868
|
Serp1
|
stress-associated endoplasmic reticulum protein 1 |
| chr6_+_51662224 | 0.12 |
ENSRNOT00000060006
|
Ccdc71l
|
coiled-coil domain containing 71-like |
| chr15_+_12569235 | 0.12 |
ENSRNOT00000083787
|
Thoc7
|
THO complex 7 |
| chr7_+_124460358 | 0.12 |
ENSRNOT00000014089
|
Tspo
|
translocator protein |
| chr2_+_188688719 | 0.12 |
ENSRNOT00000081189
|
Dcst2
|
DC-STAMP domain containing 2 |
| chr4_-_170912629 | 0.12 |
ENSRNOT00000055691
|
Erp27
|
endoplasmic reticulum protein 27 |
| chr17_+_9282675 | 0.12 |
ENSRNOT00000051702
|
H2afy
|
H2A histone family, member Y |
| chr5_-_136762986 | 0.12 |
ENSRNOT00000026852
|
Artn
|
artemin |
| chr10_+_47490153 | 0.11 |
ENSRNOT00000003182
|
Aldh3a1
|
aldehyde dehydrogenase 3 family, member A1 |
| chr6_-_44361908 | 0.11 |
ENSRNOT00000009491
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr1_+_89008117 | 0.11 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr18_-_81807627 | 0.11 |
ENSRNOT00000020376
|
Timm21
|
translocase of inner mitochondrial membrane 21 |
| chr3_+_61685619 | 0.11 |
ENSRNOT00000002140
|
Hoxd1
|
homeo box D1 |
| chr19_+_20147037 | 0.11 |
ENSRNOT00000020028
|
Zfp423
|
zinc finger protein 423 |
| chr12_+_25561777 | 0.11 |
ENSRNOT00000002032
|
Rcc1l
|
RCC1 like |
| chr10_-_90999506 | 0.11 |
ENSRNOT00000034401
|
Gfap
|
glial fibrillary acidic protein |
| chr16_+_49462889 | 0.11 |
ENSRNOT00000039909
|
Ankrd37
|
ankyrin repeat domain 37 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pparg_Rxrg
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 3.2 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.4 | 1.8 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.3 | 1.0 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.2 | 0.6 | GO:0050828 | regulation of liquid surface tension(GO:0050828) |
| 0.2 | 0.6 | GO:0030186 | melatonin metabolic process(GO:0030186) |
| 0.2 | 0.7 | GO:0009972 | cytidine catabolic process(GO:0006216) cytidine deamination(GO:0009972) cytidine metabolic process(GO:0046087) |
| 0.2 | 0.5 | GO:0043133 | hindgut contraction(GO:0043133) regulation of hindgut contraction(GO:0043134) |
| 0.2 | 0.5 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.2 | 0.2 | GO:0051542 | elastin biosynthetic process(GO:0051542) |
| 0.1 | 0.4 | GO:1990792 | response to glial cell derived neurotrophic factor(GO:1990790) cellular response to glial cell derived neurotrophic factor(GO:1990792) |
| 0.1 | 0.9 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.1 | 0.4 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.1 | 0.3 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.1 | 0.3 | GO:1904373 | response to kainic acid(GO:1904373) |
| 0.1 | 0.4 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 0.7 | GO:0030321 | transepithelial chloride transport(GO:0030321) |
| 0.1 | 0.3 | GO:0015920 | leukocyte chemotaxis involved in inflammatory response(GO:0002232) lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.9 | GO:0015671 | oxygen transport(GO:0015671) |
| 0.1 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.1 | 0.5 | GO:0035338 | fatty acid elongation, monounsaturated fatty acid(GO:0034625) long-chain fatty-acyl-CoA biosynthetic process(GO:0035338) |
| 0.1 | 0.2 | GO:1903797 | positive regulation of inorganic anion transmembrane transport(GO:1903797) |
| 0.1 | 0.2 | GO:0061017 | regulation of pronephros size(GO:0035565) pronephric nephron tubule development(GO:0039020) hepatoblast differentiation(GO:0061017) |
| 0.1 | 0.3 | GO:0042126 | nitrate metabolic process(GO:0042126) |
| 0.1 | 0.2 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.1 | 0.2 | GO:1903769 | negative regulation of cell proliferation in bone marrow(GO:1903769) |
| 0.1 | 0.2 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.1 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.1 | 0.4 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.1 | 0.2 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.1 | 0.2 | GO:0034769 | basement membrane disassembly(GO:0034769) |
| 0.1 | 0.2 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.1 | 0.4 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.1 | 0.3 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.1 | 0.2 | GO:0036112 | medium-chain fatty-acyl-CoA metabolic process(GO:0036112) |
| 0.1 | 0.2 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.1 | 0.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.1 | 0.2 | GO:0070814 | hydrogen sulfide biosynthetic process(GO:0070814) |
| 0.1 | 0.2 | GO:1900756 | protein processing in phagocytic vesicle(GO:1900756) regulation of establishment of T cell polarity(GO:1903903) regulation of protein processing in phagocytic vesicle(GO:1903921) positive regulation of protein processing in phagocytic vesicle(GO:1903923) |
| 0.0 | 0.2 | GO:0034760 | negative regulation of iron ion transport(GO:0034757) negative regulation of iron ion transmembrane transport(GO:0034760) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.1 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.0 | 0.1 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.2 | GO:0015746 | citrate transport(GO:0015746) |
| 0.0 | 0.3 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.2 | GO:0060112 | generation of ovulation cycle rhythm(GO:0060112) |
| 0.0 | 0.2 | GO:1902219 | negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress(GO:1902219) |
| 0.0 | 0.1 | GO:0015879 | carnitine transport(GO:0015879) |
| 0.0 | 0.2 | GO:0001712 | ectodermal cell fate commitment(GO:0001712) |
| 0.0 | 0.1 | GO:0033128 | negative regulation of histone phosphorylation(GO:0033128) negative regulation of protein localization to chromosome, telomeric region(GO:1904815) |
| 0.0 | 0.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.0 | 0.1 | GO:0010625 | positive regulation of Schwann cell proliferation(GO:0010625) |
| 0.0 | 0.3 | GO:0006735 | NADH regeneration(GO:0006735) canonical glycolysis(GO:0061621) glucose catabolic process to pyruvate(GO:0061718) |
| 0.0 | 0.1 | GO:0034696 | response to prostaglandin F(GO:0034696) ketone body biosynthetic process(GO:0046951) |
| 0.0 | 0.1 | GO:0070541 | response to platinum ion(GO:0070541) |
| 0.0 | 0.0 | GO:0071422 | succinate transport(GO:0015744) succinate transmembrane transport(GO:0071422) |
| 0.0 | 0.1 | GO:0071348 | cellular response to interleukin-11(GO:0071348) |
| 0.0 | 0.2 | GO:1990086 | lens fiber cell apoptotic process(GO:1990086) |
| 0.0 | 0.1 | GO:0001306 | age-dependent response to oxidative stress(GO:0001306) age-dependent general metabolic decline(GO:0007571) |
| 0.0 | 0.2 | GO:0035934 | corticosterone secretion(GO:0035934) |
| 0.0 | 0.2 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.1 | GO:0061732 | mitochondrial acetyl-CoA biosynthetic process from pyruvate(GO:0061732) |
| 0.0 | 0.1 | GO:0002477 | antigen processing and presentation of exogenous peptide antigen via MHC class Ib(GO:0002477) antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent(GO:0002481) |
| 0.0 | 0.2 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0046016 | positive regulation of transcription by glucose(GO:0046016) |
| 0.0 | 0.1 | GO:0019364 | pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.2 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0070086 | ubiquitin-dependent endocytosis(GO:0070086) |
| 0.0 | 0.2 | GO:0060762 | regulation of branching involved in mammary gland duct morphogenesis(GO:0060762) |
| 0.0 | 0.3 | GO:0034063 | stress granule assembly(GO:0034063) |
| 0.0 | 0.1 | GO:0035617 | stress granule disassembly(GO:0035617) |
| 0.0 | 0.1 | GO:0046061 | DNA protection(GO:0042262) dATP catabolic process(GO:0046061) |
| 0.0 | 0.1 | GO:0072249 | negative regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045715) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.0 | 0.2 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:1902847 | regulation of neuronal signal transduction(GO:1902847) positive regulation of tau-protein kinase activity(GO:1902949) neurofibrillary tangle assembly(GO:1902988) regulation of neurofibrillary tangle assembly(GO:1902996) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.0 | 0.3 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.2 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.4 | GO:0030150 | protein import into mitochondrial matrix(GO:0030150) |
| 0.0 | 0.1 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.2 | GO:0006559 | L-phenylalanine catabolic process(GO:0006559) erythrose 4-phosphate/phosphoenolpyruvate family amino acid catabolic process(GO:1902222) |
| 0.0 | 0.3 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.1 | GO:1990168 | protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.1 | GO:0006554 | lysine catabolic process(GO:0006554) L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0046351 | disaccharide biosynthetic process(GO:0046351) |
| 0.0 | 0.1 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.0 | GO:0007603 | phototransduction, visible light(GO:0007603) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.1 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:1990402 | embryonic liver development(GO:1990402) |
| 0.0 | 0.1 | GO:0003166 | bundle of His development(GO:0003166) negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.2 | GO:0042760 | very long-chain fatty acid catabolic process(GO:0042760) |
| 0.0 | 0.1 | GO:0015938 | coenzyme A catabolic process(GO:0015938) nucleoside bisphosphate catabolic process(GO:0033869) ribonucleoside bisphosphate catabolic process(GO:0034031) purine nucleoside bisphosphate catabolic process(GO:0034034) acetyl-CoA catabolic process(GO:0046356) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0038190 | neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) cardiac vascular smooth muscle cell development(GO:0060948) positive regulation of lymphangiogenesis(GO:1901492) |
| 0.0 | 0.0 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0006214 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 0.0 | 0.1 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.0 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.4 | GO:0042438 | melanin biosynthetic process(GO:0042438) |
| 0.0 | 0.1 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.1 | GO:0006598 | polyamine catabolic process(GO:0006598) |
| 0.0 | 0.1 | GO:0072656 | response to vitamin B1(GO:0010266) positive regulation of necrotic cell death(GO:0010940) maintenance of protein location in mitochondrion(GO:0072656) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0034971 | histone H3-R17 methylation(GO:0034971) |
| 0.0 | 0.0 | GO:0060584 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:1900620 | acetylcholine biosynthetic process(GO:0008292) acetate ester biosynthetic process(GO:1900620) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:1903659 | regulation of complement-dependent cytotoxicity(GO:1903659) |
| 0.0 | 0.1 | GO:0031446 | regulation of fast-twitch skeletal muscle fiber contraction(GO:0031446) positive regulation of fast-twitch skeletal muscle fiber contraction(GO:0031448) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.1 | GO:0006117 | acetaldehyde metabolic process(GO:0006117) |
| 0.0 | 0.1 | GO:0035470 | positive regulation of vascular wound healing(GO:0035470) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0030388 | fructose 1,6-bisphosphate metabolic process(GO:0030388) |
| 0.0 | 0.0 | GO:0021699 | cerebellar cortex maturation(GO:0021699) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.2 | GO:1901663 | ubiquinone biosynthetic process(GO:0006744) quinone biosynthetic process(GO:1901663) |
| 0.0 | 0.1 | GO:0034379 | very-low-density lipoprotein particle assembly(GO:0034379) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:1901164 | negative regulation of trophoblast cell migration(GO:1901164) |
| 0.0 | 0.0 | GO:0003100 | regulation of systemic arterial blood pressure by endothelin(GO:0003100) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.1 | GO:0060018 | astrocyte fate commitment(GO:0060018) |
| 0.0 | 0.0 | GO:0060468 | negative regulation of fertilization(GO:0060467) prevention of polyspermy(GO:0060468) |
| 0.0 | 0.1 | GO:0016024 | CDP-diacylglycerol biosynthetic process(GO:0016024) |
| 0.0 | 0.1 | GO:1903862 | positive regulation of oxidative phosphorylation(GO:1903862) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0015866 | ADP transport(GO:0015866) |
| 0.0 | 0.2 | GO:0032927 | male sex determination(GO:0030238) positive regulation of activin receptor signaling pathway(GO:0032927) lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.1 | GO:0070314 | G1 to G0 transition(GO:0070314) |
| 0.0 | 0.0 | GO:0045210 | negative regulation of dendritic cell cytokine production(GO:0002731) FasL biosynthetic process(GO:0045210) positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.0 | 0.0 | GO:0060661 | submandibular salivary gland formation(GO:0060661) positive regulation of hair follicle cell proliferation(GO:0071338) |
| 0.0 | 0.1 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.0 | 0.0 | GO:0046135 | pyrimidine nucleoside catabolic process(GO:0046135) |
| 0.0 | 0.3 | GO:0071803 | positive regulation of podosome assembly(GO:0071803) |
| 0.0 | 0.4 | GO:0015986 | energy coupled proton transport, down electrochemical gradient(GO:0015985) ATP synthesis coupled proton transport(GO:0015986) |
| 0.0 | 0.1 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:0034227 | tRNA thio-modification(GO:0034227) |
| 0.0 | 0.1 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.2 | GO:0036158 | outer dynein arm assembly(GO:0036158) |
| 0.0 | 0.0 | GO:0018171 | peptidyl-cysteine oxidation(GO:0018171) |
| 0.0 | 0.0 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.3 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.0 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.0 | GO:0061152 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.0 | 0.1 | GO:0009758 | carbohydrate utilization(GO:0009758) fructose transport(GO:0015755) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.1 | GO:0060372 | regulation of atrial cardiac muscle cell membrane repolarization(GO:0060372) |
| 0.0 | 0.0 | GO:0001545 | primary ovarian follicle growth(GO:0001545) |
| 0.0 | 0.1 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.0 | GO:0018916 | nitrobenzene metabolic process(GO:0018916) |
| 0.0 | 0.1 | GO:0032819 | positive regulation of natural killer cell proliferation(GO:0032819) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:0002361 | CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0002361) |
| 0.0 | 0.1 | GO:1901741 | positive regulation of myoblast fusion(GO:1901741) |
| 0.0 | 0.0 | GO:0098705 | copper ion import across plasma membrane(GO:0098705) copper ion import into cell(GO:1902861) |
| 0.0 | 0.1 | GO:0010886 | positive regulation of cholesterol storage(GO:0010886) |
| 0.0 | 0.0 | GO:0060332 | positive regulation of humoral immune response mediated by circulating immunoglobulin(GO:0002925) positive regulation of response to interferon-gamma(GO:0060332) positive regulation of interferon-gamma-mediated signaling pathway(GO:0060335) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.2 | GO:2001241 | positive regulation of extrinsic apoptotic signaling pathway in absence of ligand(GO:2001241) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.1 | 0.3 | GO:0032173 | septin collar(GO:0032173) |
| 0.1 | 0.4 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.1 | 0.3 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.3 | GO:0031085 | BLOC-3 complex(GO:0031085) |
| 0.1 | 0.2 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.1 | 0.3 | GO:1903349 | omegasome membrane(GO:1903349) omegasome(GO:1990462) |
| 0.0 | 0.3 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:1990590 | ATF1-ATF4 transcription factor complex(GO:1990590) |
| 0.0 | 0.1 | GO:1990879 | CST complex(GO:1990879) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0097453 | mesaxon(GO:0097453) ensheathing process(GO:1990015) |
| 0.0 | 0.2 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.0 | 0.3 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.2 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.6 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.0 | 0.1 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0055087 | Ski complex(GO:0055087) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 1.5 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.2 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0030478 | actin cap(GO:0030478) |
| 0.0 | 0.1 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.3 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.1 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.1 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.6 | GO:0005921 | gap junction(GO:0005921) |
| 0.0 | 0.1 | GO:0005744 | mitochondrial inner membrane presequence translocase complex(GO:0005744) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.6 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.1 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.1 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.0 | GO:1990907 | beta-catenin-TCF complex(GO:1990907) |
| 0.0 | 0.2 | GO:0030688 | preribosome, small subunit precursor(GO:0030688) |
| 0.0 | 1.8 | GO:0090575 | RNA polymerase II transcription factor complex(GO:0090575) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.0 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.1 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0035686 | sperm fibrous sheath(GO:0035686) |
| 0.0 | 0.1 | GO:0016460 | myosin II complex(GO:0016460) |
| 0.0 | 0.0 | GO:0005749 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.1 | GO:0000813 | ESCRT I complex(GO:0000813) |
| 0.0 | 0.1 | GO:0097433 | dense body(GO:0097433) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0031720 | haptoglobin binding(GO:0031720) |
| 0.1 | 0.7 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.1 | 0.4 | GO:0004167 | dopachrome isomerase activity(GO:0004167) |
| 0.1 | 0.5 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.3 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 0.6 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.1 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.1 | 0.3 | GO:0008940 | nitrate reductase activity(GO:0008940) |
| 0.1 | 0.2 | GO:0004657 | proline dehydrogenase activity(GO:0004657) |
| 0.1 | 0.2 | GO:0050429 | calcium-dependent phospholipase C activity(GO:0050429) |
| 0.1 | 0.2 | GO:0004736 | pyruvate carboxylase activity(GO:0004736) |
| 0.1 | 0.2 | GO:0005128 | erythropoietin receptor binding(GO:0005128) |
| 0.1 | 0.5 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.1 | 0.2 | GO:0004792 | thiosulfate sulfurtransferase activity(GO:0004792) |
| 0.1 | 0.2 | GO:0031770 | growth hormone-releasing hormone receptor binding(GO:0031770) |
| 0.1 | 0.3 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.0 | 0.3 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.0 | 0.2 | GO:0015137 | citrate transmembrane transporter activity(GO:0015137) |
| 0.0 | 0.1 | GO:0004807 | triose-phosphate isomerase activity(GO:0004807) |
| 0.0 | 0.2 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 1.8 | GO:0001102 | RNA polymerase II activating transcription factor binding(GO:0001102) |
| 0.0 | 0.1 | GO:0018479 | benzaldehyde dehydrogenase (NAD+) activity(GO:0018479) |
| 0.0 | 0.0 | GO:0016508 | long-chain-enoyl-CoA hydratase activity(GO:0016508) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0016416 | carnitine O-palmitoyltransferase activity(GO:0004095) O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0015141 | succinate transmembrane transporter activity(GO:0015141) |
| 0.0 | 0.1 | GO:0015199 | amino-acid betaine transmembrane transporter activity(GO:0015199) carnitine transmembrane transporter activity(GO:0015226) |
| 0.0 | 0.1 | GO:0036042 | long-chain fatty acyl-CoA binding(GO:0036042) |
| 0.0 | 0.2 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.2 | GO:0089720 | caspase binding(GO:0089720) |
| 0.0 | 0.1 | GO:0016822 | hydrolase activity, acting on acid carbon-carbon bonds(GO:0016822) hydrolase activity, acting on acid carbon-carbon bonds, in ketonic substances(GO:0016823) |
| 0.0 | 0.1 | GO:0016748 | succinyltransferase activity(GO:0016748) |
| 0.0 | 0.1 | GO:0004060 | arylamine N-acetyltransferase activity(GO:0004060) |
| 0.0 | 0.2 | GO:0048019 | receptor antagonist activity(GO:0048019) |
| 0.0 | 0.1 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.0 | 0.6 | GO:0008171 | O-methyltransferase activity(GO:0008171) |
| 0.0 | 0.1 | GO:0016721 | superoxide dismutase activity(GO:0004784) oxidoreductase activity, acting on superoxide radicals as acceptor(GO:0016721) |
| 0.0 | 0.6 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.2 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 1.0 | GO:0016831 | carboxy-lyase activity(GO:0016831) |
| 0.0 | 0.1 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.1 | GO:0034041 | sterol-transporting ATPase activity(GO:0034041) glycoprotein transporter activity(GO:0034437) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.0 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.1 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.0 | 0.1 | GO:0004159 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.5 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.6 | GO:0005184 | neuropeptide hormone activity(GO:0005184) |
| 0.0 | 0.1 | GO:0035375 | zymogen binding(GO:0035375) |
| 0.0 | 0.2 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0004466 | long-chain-acyl-CoA dehydrogenase activity(GO:0004466) |
| 0.0 | 0.1 | GO:0010385 | double-stranded methylated DNA binding(GO:0010385) |
| 0.0 | 0.1 | GO:0035251 | UDP-glucosyltransferase activity(GO:0035251) |
| 0.0 | 0.1 | GO:0004127 | cytidylate kinase activity(GO:0004127) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.1 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.1 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.0 | 0.1 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0032051 | clathrin light chain binding(GO:0032051) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.4 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.1 | GO:0016624 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, disulfide as acceptor(GO:0016624) |
| 0.0 | 0.4 | GO:0051537 | 2 iron, 2 sulfur cluster binding(GO:0051537) |
| 0.0 | 0.1 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.1 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0004963 | follicle-stimulating hormone receptor activity(GO:0004963) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
| 0.0 | 0.0 | GO:0031783 | corticotropin hormone receptor binding(GO:0031780) type 5 melanocortin receptor binding(GO:0031783) |
| 0.0 | 0.1 | GO:0003988 | acetyl-CoA C-acyltransferase activity(GO:0003988) |
| 0.0 | 1.1 | GO:0038024 | cargo receptor activity(GO:0038024) |
| 0.0 | 0.1 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.3 | GO:0004890 | GABA-A receptor activity(GO:0004890) |
| 0.0 | 0.2 | GO:0070008 | serine-type exopeptidase activity(GO:0070008) |
| 0.0 | 0.1 | GO:0019808 | polyamine binding(GO:0019808) |
| 0.0 | 0.1 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.0 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0038181 | bile acid receptor activity(GO:0038181) |
| 0.0 | 0.1 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.1 | GO:0043184 | vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0032427 | GBD domain binding(GO:0032427) |
| 0.0 | 0.0 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 0.1 | GO:0016803 | ether hydrolase activity(GO:0016803) |
| 0.0 | 0.9 | GO:0005254 | chloride channel activity(GO:0005254) |
| 0.0 | 0.1 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.0 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0015217 | ATP transmembrane transporter activity(GO:0005347) ADP transmembrane transporter activity(GO:0015217) |
| 0.0 | 0.2 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 0.1 | GO:1990825 | sequence-specific mRNA binding(GO:1990825) |
| 0.0 | 0.0 | GO:0008265 | Mo-molybdopterin cofactor sulfurase activity(GO:0008265) |
| 0.0 | 0.1 | GO:0034185 | apolipoprotein binding(GO:0034185) |
| 0.0 | 0.1 | GO:0043522 | leucine zipper domain binding(GO:0043522) |
| 0.0 | 0.2 | GO:0043176 | amine binding(GO:0043176) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.2 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.0 | GO:0061665 | SUMO ligase activity(GO:0061665) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.3 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.2 | GO:0019203 | carbohydrate phosphatase activity(GO:0019203) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID FOXO PATHWAY | FoxO family signaling |
| 0.0 | 0.2 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 0.1 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.8 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.4 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.7 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.9 | REACTOME SULFUR AMINO ACID METABOLISM | Genes involved in Sulfur amino acid metabolism |
| 0.0 | 0.7 | REACTOME FATTY ACYL COA BIOSYNTHESIS | Genes involved in Fatty Acyl-CoA Biosynthesis |
| 0.0 | 0.2 | REACTOME REGULATION OF BETA CELL DEVELOPMENT | Genes involved in Regulation of beta-cell development |
| 0.0 | 0.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.8 | REACTOME GLUCONEOGENESIS | Genes involved in Gluconeogenesis |
| 0.0 | 0.5 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 0.1 | REACTOME MRNA PROCESSING | Genes involved in mRNA Processing |
| 0.0 | 0.1 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.3 | REACTOME ASSOCIATION OF LICENSING FACTORS WITH THE PRE REPLICATIVE COMPLEX | Genes involved in Association of licensing factors with the pre-replicative complex |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |
| 0.0 | 0.2 | REACTOME ADVANCED GLYCOSYLATION ENDPRODUCT RECEPTOR SIGNALING | Genes involved in Advanced glycosylation endproduct receptor signaling |
| 0.0 | 0.2 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME ALPHA LINOLENIC ACID ALA METABOLISM | Genes involved in alpha-linolenic acid (ALA) metabolism |
| 0.0 | 0.2 | REACTOME FORMATION OF ATP BY CHEMIOSMOTIC COUPLING | Genes involved in Formation of ATP by chemiosmotic coupling |
| 0.0 | 0.2 | REACTOME POST CHAPERONIN TUBULIN FOLDING PATHWAY | Genes involved in Post-chaperonin tubulin folding pathway |
| 0.0 | 0.1 | REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | Genes involved in SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs |
| 0.0 | 0.3 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.0 | 0.1 | REACTOME REGULATED PROTEOLYSIS OF P75NTR | Genes involved in Regulated proteolysis of p75NTR |
| 0.0 | 0.2 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.2 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.2 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME LIPOPROTEIN METABOLISM | Genes involved in Lipoprotein metabolism |