Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
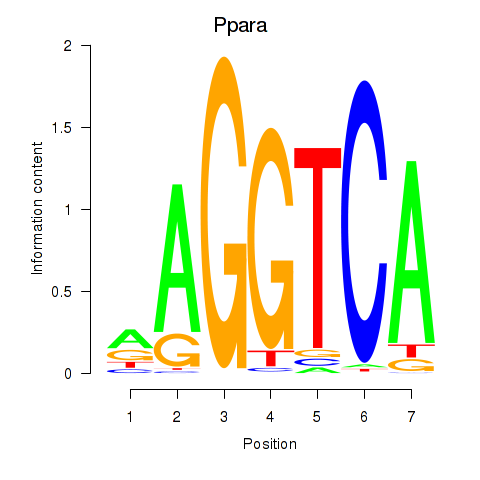
Results for Ppara
Z-value: 0.59
Transcription factors associated with Ppara
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Ppara
|
ENSRNOG00000021463 | peroxisome proliferator activated receptor alpha |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Ppara | rn6_v1_chr7_+_126619196_126619196 | 0.59 | 3.0e-01 | Click! |
Activity profile of Ppara motif
Sorted Z-values of Ppara motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_-_34550850 | 0.48 |
ENSRNOT00000027794
ENSRNOT00000090228 |
Cbln3
|
cerebellin 3 precursor |
| chr20_+_3827367 | 0.37 |
ENSRNOT00000079967
|
Rxrb
|
retinoid X receptor beta |
| chr7_+_121311024 | 0.37 |
ENSRNOT00000092260
ENSRNOT00000023066 ENSRNOT00000081377 |
Syngr1
|
synaptogyrin 1 |
| chr2_+_196334626 | 0.30 |
ENSRNOT00000050914
ENSRNOT00000028645 ENSRNOT00000090729 |
Sema6c
|
semaphorin 6C |
| chr8_-_45215974 | 0.30 |
ENSRNOT00000010792
|
Crtam
|
cytotoxic and regulatory T cell molecule |
| chr6_+_28515025 | 0.28 |
ENSRNOT00000088033
ENSRNOT00000005317 ENSRNOT00000081141 |
Dnajc27
|
DnaJ heat shock protein family (Hsp40) member C27 |
| chr4_+_51614676 | 0.28 |
ENSRNOT00000060494
|
Asb15
|
ankyrin repeat and SOCS box containing 15 |
| chr1_+_274245184 | 0.28 |
ENSRNOT00000018889
|
Dusp5
|
dual specificity phosphatase 5 |
| chr10_-_71849293 | 0.27 |
ENSRNOT00000003799
|
Lhx1
|
LIM homeobox 1 |
| chr4_+_8256611 | 0.23 |
ENSRNOT00000061894
|
AABR07059198.1
|
|
| chr3_+_131351587 | 0.22 |
ENSRNOT00000010835
|
Btbd3
|
BTB domain containing 3 |
| chr1_+_197659187 | 0.22 |
ENSRNOT00000082228
|
LOC103690016
|
serine/threonine-protein kinase SBK1 |
| chr11_+_61531416 | 0.21 |
ENSRNOT00000093263
|
Atp6v1a
|
ATPase H+ transporting V1 subunit A |
| chr4_+_78263866 | 0.21 |
ENSRNOT00000033807
|
AI854703
|
expressed sequence AI854703 |
| chr3_+_147609095 | 0.20 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr20_+_3351303 | 0.20 |
ENSRNOT00000080419
ENSRNOT00000001065 ENSRNOT00000086503 |
Atat1
|
alpha tubulin acetyltransferase 1 |
| chr14_-_83741969 | 0.20 |
ENSRNOT00000026293
|
Inpp5j
|
inositol polyphosphate-5-phosphatase J |
| chr2_-_123396147 | 0.19 |
ENSRNOT00000079004
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr10_-_5196892 | 0.19 |
ENSRNOT00000083982
|
Clec16a
|
C-type lectin domain family 16, member A |
| chr6_-_106971250 | 0.18 |
ENSRNOT00000010926
|
Dpf3
|
double PHD fingers 3 |
| chr1_-_201906286 | 0.18 |
ENSRNOT00000064511
ENSRNOT00000036625 |
RGD1560958
|
similar to RIKEN cDNA 1700063I17 |
| chr1_+_82174451 | 0.17 |
ENSRNOT00000027783
|
Tmem145
|
transmembrane protein 145 |
| chrX_-_40086870 | 0.17 |
ENSRNOT00000010027
|
Smpx
|
small muscle protein, X-linked |
| chr7_-_127081704 | 0.17 |
ENSRNOT00000055893
|
LOC108351584
|
uncharacterized LOC108351584 |
| chr8_+_59900651 | 0.17 |
ENSRNOT00000020410
|
Tmem266
|
transmembrane protein 266 |
| chr12_-_44520341 | 0.17 |
ENSRNOT00000066810
|
Nos1
|
nitric oxide synthase 1 |
| chr5_-_152473868 | 0.16 |
ENSRNOT00000022130
|
Fam110d
|
family with sequence similarity 110, member D |
| chr8_+_116332796 | 0.16 |
ENSRNOT00000021408
|
Hyal1
|
hyaluronoglucosaminidase 1 |
| chr13_+_47440803 | 0.16 |
ENSRNOT00000030476
|
Yod1
|
YOD1 deubiquitinase |
| chr10_-_85517683 | 0.16 |
ENSRNOT00000016070
|
Srcin1
|
SRC kinase signaling inhibitor 1 |
| chr3_+_62648447 | 0.16 |
ENSRNOT00000002111
|
Agps
|
alkylglycerone phosphate synthase |
| chr13_+_110398800 | 0.16 |
ENSRNOT00000064492
|
Lpgat1
|
lysophosphatidylglycerol acyltransferase 1 |
| chr7_-_125465848 | 0.16 |
ENSRNOT00000039588
|
Ldoc1l
|
leucine zipper, down-regulated in cancer 1-like |
| chr20_-_5618254 | 0.15 |
ENSRNOT00000092326
ENSRNOT00000000576 |
Bak1
|
BCL2-antagonist/killer 1 |
| chr4_-_100232559 | 0.15 |
ENSRNOT00000016984
|
Vamp5
|
vesicle-associated membrane protein 5 |
| chr10_-_90940118 | 0.15 |
ENSRNOT00000093143
|
Eftud2
|
elongation factor Tu GTP binding domain containing 2 |
| chr3_+_95715193 | 0.15 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chr5_-_75319765 | 0.15 |
ENSRNOT00000085698
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr13_-_88642011 | 0.15 |
ENSRNOT00000067037
|
LOC100361087
|
hypothetical LOC100361087 |
| chr5_+_147069616 | 0.15 |
ENSRNOT00000072908
|
Trim62
|
tripartite motif-containing 62 |
| chr8_+_116094851 | 0.15 |
ENSRNOT00000084120
|
RGD1307461
|
similar to RIKEN cDNA 6430571L13 gene; similar to g20 protein |
| chr2_+_186980992 | 0.14 |
ENSRNOT00000020717
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr18_-_26211445 | 0.14 |
ENSRNOT00000027739
|
Nrep
|
neuronal regeneration related protein |
| chr10_+_56381813 | 0.14 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr6_+_110624856 | 0.14 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr18_+_73564247 | 0.14 |
ENSRNOT00000077679
ENSRNOT00000035317 |
St8sia5
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 5 |
| chr7_-_12424367 | 0.14 |
ENSRNOT00000060698
|
Midn
|
midnolin |
| chr8_-_62248013 | 0.13 |
ENSRNOT00000080012
ENSRNOT00000089602 |
Scamp5
|
secretory carrier membrane protein 5 |
| chr12_-_2555164 | 0.13 |
ENSRNOT00000084460
ENSRNOT00000061821 |
Map2k7
|
mitogen activated protein kinase kinase 7 |
| chr1_-_49844547 | 0.13 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr2_-_174413236 | 0.13 |
ENSRNOT00000064588
|
Golim4
|
golgi integral membrane protein 4 |
| chr9_+_10471742 | 0.13 |
ENSRNOT00000072276
|
Safb2
|
scaffold attachment factor B2 |
| chr7_+_127081978 | 0.13 |
ENSRNOT00000022945
|
Tbc1d22a
|
TBC1 domain family, member 22a |
| chr3_-_7795758 | 0.12 |
ENSRNOT00000045919
|
Ntng2
|
netrin G2 |
| chr10_-_91448477 | 0.12 |
ENSRNOT00000038836
|
Arhgap27
|
Rho GTPase activating protein 27 |
| chr3_+_177226417 | 0.12 |
ENSRNOT00000031328
|
Oprl1
|
opioid related nociceptin receptor 1 |
| chr10_-_90995982 | 0.12 |
ENSRNOT00000093266
|
Gfap
|
glial fibrillary acidic protein |
| chr3_-_72113680 | 0.12 |
ENSRNOT00000009708
|
Zdhhc5
|
zinc finger, DHHC-type containing 5 |
| chr12_+_21767606 | 0.12 |
ENSRNOT00000059602
|
LOC103691013
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr6_+_104340206 | 0.12 |
ENSRNOT00000058301
|
Plekhd1
|
pleckstrin homology and coiled-coil domain containing D1 |
| chr10_-_63952726 | 0.12 |
ENSRNOT00000090461
|
Doc2b
|
double C2 domain beta |
| chr3_-_164239250 | 0.12 |
ENSRNOT00000012604
|
Spata2
|
spermatogenesis associated 2 |
| chrX_-_123662350 | 0.11 |
ENSRNOT00000092624
|
Sept6
|
septin 6 |
| chr2_+_186980793 | 0.11 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr3_+_151126591 | 0.11 |
ENSRNOT00000025859
|
Myh7b
|
myosin heavy chain 7B |
| chr19_-_56789404 | 0.11 |
ENSRNOT00000058652
|
RGD1563991
|
similar to TAF5-like RNA polymerase II, p300/CBP-associated factor (PCAF)-associated factor |
| chr4_+_153385205 | 0.11 |
ENSRNOT00000016561
|
Bcl2l13
|
BCL2 like 13 |
| chr6_+_26390689 | 0.11 |
ENSRNOT00000079762
ENSRNOT00000091528 |
Ift172
|
intraflagellar transport 172 |
| chr8_+_117280705 | 0.11 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr6_+_146784915 | 0.11 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr2_+_45668969 | 0.11 |
ENSRNOT00000014761
ENSRNOT00000071353 |
Arl15
|
ADP-ribosylation factor like GTPase 15 |
| chr4_+_119815139 | 0.11 |
ENSRNOT00000083402
ENSRNOT00000016137 |
Hmces
|
5-hydroxymethylcytosine (hmC) binding, ES cell-specific |
| chr10_+_88764732 | 0.11 |
ENSRNOT00000026662
|
Stat5a
|
signal transducer and activator of transcription 5A |
| chr8_-_46750898 | 0.11 |
ENSRNOT00000059951
|
Tbcel
|
tubulin folding cofactor E-like |
| chr5_-_147148291 | 0.10 |
ENSRNOT00000085151
|
Azin2
|
antizyme inhibitor 2 |
| chr4_+_68656928 | 0.10 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr10_+_61432819 | 0.10 |
ENSRNOT00000003687
ENSRNOT00000092478 |
Cluh
|
clustered mitochondria homolog |
| chr1_+_184271590 | 0.10 |
ENSRNOT00000014764
|
Calcb
|
calcitonin-related polypeptide, beta |
| chr7_+_144052061 | 0.10 |
ENSRNOT00000020103
ENSRNOT00000091378 |
Amhr2
|
anti-Mullerian hormone receptor type 2 |
| chr14_+_104191517 | 0.10 |
ENSRNOT00000006573
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr13_+_110920830 | 0.10 |
ENSRNOT00000077014
ENSRNOT00000076362 |
Kcnh1
|
potassium voltage-gated channel subfamily H member 1 |
| chr7_-_120518653 | 0.10 |
ENSRNOT00000016362
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr1_+_167374182 | 0.10 |
ENSRNOT00000092882
|
Stim1
|
stromal interaction molecule 1 |
| chr7_+_141326950 | 0.10 |
ENSRNOT00000084075
|
Asic1
|
acid sensing ion channel subunit 1 |
| chr1_-_101175570 | 0.10 |
ENSRNOT00000073918
|
Gfy
|
golgi-associated, olfactory signaling regulator |
| chr10_-_15311189 | 0.10 |
ENSRNOT00000027371
|
Prr35
|
proline rich 35 |
| chr1_-_88826302 | 0.09 |
ENSRNOT00000028275
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr12_-_12907771 | 0.09 |
ENSRNOT00000001412
|
Cyth3
|
cytohesin 3 |
| chr9_-_10471009 | 0.09 |
ENSRNOT00000072868
|
Safb
|
scaffold attachment factor B |
| chr1_+_79831534 | 0.09 |
ENSRNOT00000057965
|
Nova2
|
NOVA alternative splicing regulator 2 |
| chr1_-_7480825 | 0.09 |
ENSRNOT00000048754
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr2_-_118745766 | 0.09 |
ENSRNOT00000013858
|
Zmat3
|
zinc finger, matrin type 3 |
| chr8_+_98745310 | 0.09 |
ENSRNOT00000019938
|
Zic4
|
Zic family member 4 |
| chr10_+_75055020 | 0.09 |
ENSRNOT00000010755
|
Tspoap1
|
TSPO associated protein 1 |
| chrX_-_159841072 | 0.09 |
ENSRNOT00000001158
|
Arhgef6
|
Rac/Cdc42 guanine nucleotide exchange factor 6 |
| chr4_-_123713319 | 0.09 |
ENSRNOT00000012875
ENSRNOT00000086982 |
Slc6a6
|
solute carrier family 6 member 6 |
| chr18_-_55891710 | 0.09 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr10_-_82399484 | 0.08 |
ENSRNOT00000082972
ENSRNOT00000055582 |
Xylt2
|
xylosyltransferase 2 |
| chr10_+_27973681 | 0.08 |
ENSRNOT00000004970
|
Gabrb2
|
gamma-aminobutyric acid type A receptor beta 2 subunit |
| chr2_-_119140110 | 0.08 |
ENSRNOT00000058810
|
AABR07009978.1
|
|
| chr12_-_30033357 | 0.08 |
ENSRNOT00000001198
|
Kctd7
|
potassium channel tetramerization domain containing 7 |
| chr14_+_85230648 | 0.08 |
ENSRNOT00000089866
|
Ap1b1
|
adaptor-related protein complex 1, beta 1 subunit |
| chr14_-_114649173 | 0.08 |
ENSRNOT00000083528
|
Sptbn1
|
spectrin, beta, non-erythrocytic 1 |
| chr3_+_93495106 | 0.08 |
ENSRNOT00000029922
ENSRNOT00000085760 |
Abtb2
|
ankyrin repeat and BTB domain containing 2 |
| chr10_+_47019326 | 0.08 |
ENSRNOT00000006984
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr8_-_47339343 | 0.08 |
ENSRNOT00000081007
|
Arhgef12
|
Rho guanine nucleotide exchange factor 12 |
| chr12_+_40018937 | 0.08 |
ENSRNOT00000001697
|
Cux2
|
cut-like homeobox 2 |
| chr1_-_250630677 | 0.08 |
ENSRNOT00000016688
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr16_+_1979191 | 0.08 |
ENSRNOT00000014382
|
Ppif
|
peptidylprolyl isomerase F |
| chr1_-_221041401 | 0.08 |
ENSRNOT00000064136
|
Pcnx3
|
pecanex homolog 3 (Drosophila) |
| chr4_-_77706994 | 0.08 |
ENSRNOT00000038517
|
Zfp777
|
zinc finger protein 777 |
| chr4_-_89281222 | 0.08 |
ENSRNOT00000010898
|
Fam13a
|
family with sequence similarity 13, member A |
| chr17_+_15845931 | 0.08 |
ENSRNOT00000092083
|
Card19
|
caspase recruitment domain family, member 19 |
| chr10_+_47018974 | 0.08 |
ENSRNOT00000079375
|
Smcr8
|
Smith-Magenis syndrome chromosome region, candidate 8 |
| chr10_-_10881844 | 0.08 |
ENSRNOT00000087118
ENSRNOT00000004416 |
Mgrn1
|
mahogunin ring finger 1 |
| chr1_+_219144205 | 0.08 |
ENSRNOT00000083942
|
Unc93b1
|
unc-93 homolog B1 (C. elegans) |
| chr10_-_104748003 | 0.08 |
ENSRNOT00000042372
ENSRNOT00000046754 |
Acox1
|
acyl-CoA oxidase 1 |
| chr10_-_94460732 | 0.08 |
ENSRNOT00000014508
|
Smarcd2
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 2 |
| chr5_-_138545404 | 0.08 |
ENSRNOT00000011586
|
Rimkla
|
ribosomal modification protein rimK-like family member A |
| chr6_+_106496992 | 0.08 |
ENSRNOT00000086136
ENSRNOT00000058181 |
Rgs6
|
regulator of G-protein signaling 6 |
| chr3_+_149935731 | 0.08 |
ENSRNOT00000021907
|
Cbfa2t2
|
CBFA2/RUNX1 translocation partner 2 |
| chr2_+_208420206 | 0.07 |
ENSRNOT00000090500
ENSRNOT00000076065 ENSRNOT00000073644 ENSRNOT00000086279 |
LOC100911453
Wdr77
|
methylosome protein 50-like WD repeat domain 77 |
| chr3_+_80676820 | 0.07 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr16_-_19996716 | 0.07 |
ENSRNOT00000024434
ENSRNOT00000080706 |
Nxnl1
|
nucleoredoxin-like 1 |
| chr4_+_64088900 | 0.07 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr1_+_88182585 | 0.07 |
ENSRNOT00000044145
|
Yif1b
|
Yip1 interacting factor homolog B, membrane trafficking protein |
| chr17_-_48562838 | 0.07 |
ENSRNOT00000017102
ENSRNOT00000084702 |
Amph
|
amphiphysin |
| chr17_-_10952441 | 0.07 |
ENSRNOT00000024580
|
Hrh2
|
histamine receptor H 2 |
| chr1_+_145770135 | 0.07 |
ENSRNOT00000015320
|
Stard5
|
StAR-related lipid transfer domain containing 5 |
| chr18_+_28653592 | 0.07 |
ENSRNOT00000085120
ENSRNOT00000045795 |
Cxxc5
|
CXXC finger protein 5 |
| chr3_+_122114108 | 0.07 |
ENSRNOT00000091935
|
Sirpa
|
signal-regulatory protein alpha |
| chr10_-_49009156 | 0.07 |
ENSRNOT00000072970
|
Mapk7
|
mitogen-activated protein kinase 7 |
| chr6_-_76552559 | 0.07 |
ENSRNOT00000065230
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr7_-_81592206 | 0.07 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr3_+_80556668 | 0.07 |
ENSRNOT00000079118
|
Arhgap1
|
Rho GTPase activating protein 1 |
| chr4_-_82173207 | 0.07 |
ENSRNOT00000074167
|
Hoxa5
|
homeo box A5 |
| chr1_+_220428481 | 0.07 |
ENSRNOT00000027335
|
LOC108348044
|
ras and Rab interactor 1 |
| chr19_-_26053762 | 0.07 |
ENSRNOT00000004646
|
MAST1
|
microtubule associated serine/threonine kinase 1 |
| chr5_+_147692391 | 0.07 |
ENSRNOT00000064855
|
Fam229a
|
family with sequence similarity 229, member A |
| chr15_-_56970365 | 0.07 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr5_-_137372524 | 0.07 |
ENSRNOT00000009061
|
Tmem125
|
transmembrane protein 125 |
| chr20_+_28572242 | 0.07 |
ENSRNOT00000072485
|
Sh3rf3
|
SH3 domain containing ring finger 3 |
| chr10_-_65066612 | 0.07 |
ENSRNOT00000043448
|
Myo18a
|
myosin XVIIIa |
| chr2_-_123396386 | 0.07 |
ENSRNOT00000046700
|
Trpc3
|
transient receptor potential cation channel, subfamily C, member 3 |
| chr5_-_57632177 | 0.07 |
ENSRNOT00000080787
ENSRNOT00000092581 |
Ubap2
|
ubiquitin-associated protein 2 |
| chr1_+_141767940 | 0.06 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr1_+_31264755 | 0.06 |
ENSRNOT00000028988
|
LOC679739
|
NADH dehydrogenase (ubiquinone) Fe-S protein 6 |
| chr6_+_12415805 | 0.06 |
ENSRNOT00000022380
|
Gtf2a1l
|
general transcription factor 2A subunit 1 like |
| chr7_-_12311149 | 0.06 |
ENSRNOT00000046886
|
Dazap1
|
DAZ associated protein 1 |
| chr13_-_89306219 | 0.06 |
ENSRNOT00000004183
ENSRNOT00000079247 |
Fcrla
|
Fc receptor-like A |
| chr12_+_24761210 | 0.06 |
ENSRNOT00000002003
|
Cldn4
|
claudin 4 |
| chr9_-_92291220 | 0.06 |
ENSRNOT00000093357
|
Dner
|
delta/notch-like EGF repeat containing |
| chr14_-_85318194 | 0.06 |
ENSRNOT00000065135
|
Gas2l1
|
growth arrest-specific 2 like 1 |
| chr5_-_166695134 | 0.06 |
ENSRNOT00000047763
|
Tmem201
|
transmembrane protein 201 |
| chr6_+_147876237 | 0.06 |
ENSRNOT00000056649
|
Tmem196
|
transmembrane protein 196 |
| chr10_-_86688730 | 0.06 |
ENSRNOT00000055333
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr1_-_175586802 | 0.06 |
ENSRNOT00000071333
|
AABR07005027.1
|
|
| chr13_-_47831110 | 0.06 |
ENSRNOT00000061070
|
Mapkapk2
|
mitogen-activated protein kinase-activated protein kinase 2 |
| chr8_+_114867062 | 0.06 |
ENSRNOT00000074771
|
Wdr82
|
WD repeat domain 82 |
| chr15_-_33285779 | 0.06 |
ENSRNOT00000036150
|
Cdh24
|
cadherin 24 |
| chr6_-_107678156 | 0.06 |
ENSRNOT00000014158
|
Elmsan1
|
ELM2 and Myb/SANT domain containing 1 |
| chr1_+_266866931 | 0.06 |
ENSRNOT00000027512
|
Pdcd11
|
programmed cell death 11 |
| chr5_-_22799090 | 0.06 |
ENSRNOT00000076426
ENSRNOT00000077044 |
Asph
|
aspartate-beta-hydroxylase |
| chr3_+_48096954 | 0.06 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chr4_-_119927705 | 0.06 |
ENSRNOT00000077624
|
Rab7a
|
RAB7A, member RAS oncogene family |
| chr16_-_20460959 | 0.06 |
ENSRNOT00000026457
|
Pde4c
|
phosphodiesterase 4C |
| chr7_-_14351918 | 0.06 |
ENSRNOT00000008827
|
Akap8
|
A-kinase anchoring protein 8 |
| chr12_+_38368693 | 0.06 |
ENSRNOT00000083261
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr10_-_27179900 | 0.06 |
ENSRNOT00000082445
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr14_+_83724933 | 0.06 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr8_+_109455786 | 0.06 |
ENSRNOT00000039593
|
Msl2
|
male-specific lethal 2 homolog (Drosophila) |
| chr1_+_224882439 | 0.06 |
ENSRNOT00000024785
|
Chrm1
|
cholinergic receptor, muscarinic 1 |
| chr10_-_40375605 | 0.06 |
ENSRNOT00000014464
|
Anxa6
|
annexin A6 |
| chrX_+_53360839 | 0.06 |
ENSRNOT00000091467
ENSRNOT00000034372 ENSRNOT00000081061 |
Dmd
|
dystrophin |
| chr8_+_29453643 | 0.06 |
ENSRNOT00000090643
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr1_-_88066101 | 0.06 |
ENSRNOT00000079473
ENSRNOT00000027893 |
Ryr1
|
ryanodine receptor 1 |
| chr1_+_87180624 | 0.06 |
ENSRNOT00000056933
ENSRNOT00000027987 |
LOC103689986
|
protein YIF1B |
| chr13_-_25262469 | 0.05 |
ENSRNOT00000019921
|
Rnf152
|
ring finger protein 152 |
| chr9_-_9702306 | 0.05 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr9_-_71830730 | 0.05 |
ENSRNOT00000019963
|
Cryga
|
crystallin, gamma A |
| chr4_-_130659697 | 0.05 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chrX_-_113584459 | 0.05 |
ENSRNOT00000025923
|
Kcne5
|
potassium voltage-gated channel subfamily E regulatory subunit 5 |
| chr8_+_36125999 | 0.05 |
ENSRNOT00000013036
ENSRNOT00000088046 |
Kirrel3
|
kin of IRRE like 3 (Drosophila) |
| chr6_+_52663112 | 0.05 |
ENSRNOT00000013842
|
Atxn7l1
|
ataxin 7-like 1 |
| chr4_+_49941304 | 0.05 |
ENSRNOT00000008719
|
Ptprz1
|
protein tyrosine phosphatase, receptor type Z1 |
| chr1_-_204805738 | 0.05 |
ENSRNOT00000086830
|
Fam53b
|
family with sequence similarity 53, member B |
| chr15_-_106678918 | 0.05 |
ENSRNOT00000034723
|
Stk24
|
serine/threonine kinase 24 |
| chr5_+_154522119 | 0.05 |
ENSRNOT00000072618
|
E2f2
|
E2F transcription factor 2 |
| chr14_+_104821215 | 0.05 |
ENSRNOT00000007162
|
Sertad2
|
SERTA domain containing 2 |
| chr5_+_154552195 | 0.05 |
ENSRNOT00000072864
|
Asap3
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 |
| chr2_+_116028781 | 0.05 |
ENSRNOT00000089477
|
Phc3
|
polyhomeotic homolog 3 |
| chr3_+_163570532 | 0.05 |
ENSRNOT00000010054
|
Arfgef2
|
ADP ribosylation factor guanine nucleotide exchange factor 2 |
| chr3_+_172856733 | 0.05 |
ENSRNOT00000009826
|
Edn3
|
endothelin 3 |
| chr18_-_27749235 | 0.05 |
ENSRNOT00000026696
|
Hspa9
|
heat shock protein family A member 9 |
| chr1_+_185427736 | 0.05 |
ENSRNOT00000064706
|
Plekha7
|
pleckstrin homology domain containing A7 |
| chr1_+_222519615 | 0.05 |
ENSRNOT00000083585
|
Rcor2
|
REST corepressor 2 |
| chr4_+_123801174 | 0.05 |
ENSRNOT00000029055
|
RGD1560289
|
similar to chromosome 3 open reading frame 20 |
| chr16_+_23668595 | 0.05 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_+_116421894 | 0.05 |
ENSRNOT00000080577
ENSRNOT00000086628 ENSRNOT00000004017 |
Nfia
|
nuclear factor I/A |
| chr6_+_110093819 | 0.05 |
ENSRNOT00000013602
|
Gpatch2l
|
G patch domain containing 2-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Ppara
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060066 | metanephric part of ureteric bud development(GO:0035502) oviduct development(GO:0060066) |
| 0.1 | 0.2 | GO:0071929 | alpha-tubulin acetylation(GO:0071929) |
| 0.1 | 0.3 | GO:1903244 | positive regulation of cardiac muscle adaptation(GO:0010615) positive regulation of cardiac muscle hypertrophy in response to stress(GO:1903244) |
| 0.1 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.0 | 0.3 | GO:1901098 | positive regulation of autophagosome maturation(GO:1901098) |
| 0.0 | 0.1 | GO:0003322 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.3 | GO:0002860 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.2 | GO:1901204 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) |
| 0.0 | 0.2 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.2 | GO:0030210 | heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.1 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.0 | 0.1 | GO:1903300 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0060764 | cell-cell signaling involved in mammary gland development(GO:0060764) |
| 0.0 | 0.2 | GO:1902262 | B cell selection(GO:0002339) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in patterning of blood vessels(GO:1902262) |
| 0.0 | 0.1 | GO:1902269 | positive regulation of polyamine transmembrane transport(GO:1902269) |
| 0.0 | 0.1 | GO:1902445 | regulation of mitochondrial membrane permeability involved in programmed necrotic cell death(GO:1902445) |
| 0.0 | 0.1 | GO:0046544 | negative regulation of mast cell apoptotic process(GO:0033026) development of secondary male sexual characteristics(GO:0046544) regulation of mast cell differentiation(GO:0060375) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0033540 | fatty acid beta-oxidation using acyl-CoA oxidase(GO:0033540) |
| 0.0 | 0.2 | GO:1904714 | positive regulation of Schwann cell proliferation(GO:0010625) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.1 | GO:0001698 | gastrin-induced gastric acid secretion(GO:0001698) |
| 0.0 | 0.1 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.1 | GO:0090164 | asymmetric Golgi ribbon formation(GO:0090164) |
| 0.0 | 0.1 | GO:0007197 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:1901503 | ether lipid biosynthetic process(GO:0008611) glycerol ether biosynthetic process(GO:0046504) ether biosynthetic process(GO:1901503) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.0 | 0.1 | GO:0010901 | regulation of very-low-density lipoprotein particle remodeling(GO:0010901) |
| 0.0 | 0.0 | GO:0061441 | renal artery morphogenesis(GO:0061441) |
| 0.0 | 0.1 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.0 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.0 | GO:1902037 | negative regulation of hematopoietic stem cell differentiation(GO:1902037) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0060528 | secretory columnal luminar epithelial cell differentiation involved in prostate glandular acinus development(GO:0060528) |
| 0.0 | 0.4 | GO:0055012 | ventricular cardiac muscle cell differentiation(GO:0055012) |
| 0.0 | 0.1 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.1 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.1 | GO:1900147 | Schwann cell migration(GO:0036135) regulation of Schwann cell migration(GO:1900147) |
| 0.0 | 0.0 | GO:1900738 | positive regulation of phospholipase C-activating G-protein coupled receptor signaling pathway(GO:1900738) |
| 0.0 | 0.0 | GO:0030997 | regulation of centriole-centriole cohesion(GO:0030997) |
| 0.0 | 0.0 | GO:0009957 | epidermal cell fate specification(GO:0009957) regulation of ephrin receptor signaling pathway(GO:1901187) |
| 0.0 | 0.3 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.0 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0071420 | cellular response to histamine(GO:0071420) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.0 | GO:0015864 | pyrimidine nucleoside transport(GO:0015864) |
| 0.0 | 0.0 | GO:0048861 | oncostatin-M-mediated signaling pathway(GO:0038165) leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.0 | 0.1 | GO:2001184 | toll-like receptor 7 signaling pathway(GO:0034154) positive regulation of interleukin-12 secretion(GO:2001184) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0036117 | hyaluranon cable(GO:0036117) |
| 0.1 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.2 | GO:0098574 | cytoplasmic side of lysosomal membrane(GO:0098574) |
| 0.0 | 0.1 | GO:0032173 | septin collar(GO:0032173) |
| 0.0 | 0.1 | GO:0016514 | SWI/SNF complex(GO:0016514) |
| 0.0 | 0.2 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.1 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:0097598 | sperm cytoplasmic droplet(GO:0097598) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.0 | 0.1 | GO:0072487 | MSL complex(GO:0072487) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.2 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.1 | GO:1990316 | ATG1/ULK1 kinase complex(GO:1990316) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034485 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.1 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0030156 | benzodiazepine receptor binding(GO:0030156) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0030977 | taurine binding(GO:0030977) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0015489 | polyamine transmembrane transporter activity(GO:0015203) putrescine transmembrane transporter activity(GO:0015489) ornithine decarboxylase activator activity(GO:0042978) |
| 0.0 | 0.3 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0072590 | N-acetyl-L-aspartate-L-glutamate ligase activity(GO:0072590) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.3 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.3 | GO:0070679 | inositol 1,4,5 trisphosphate binding(GO:0070679) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.0 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003997 | acyl-CoA oxidase activity(GO:0003997) |
| 0.0 | 0.0 | GO:0004924 | oncostatin-M receptor activity(GO:0004924) |
| 0.0 | 0.1 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.0 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.0 | GO:0001716 | L-amino-acid oxidase activity(GO:0001716) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.1 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005237 | inhibitory extracellular ligand-gated ion channel activity(GO:0005237) |
| 0.0 | 0.1 | GO:0008527 | taste receptor activity(GO:0008527) |
| 0.0 | 0.0 | GO:0004655 | porphobilinogen synthase activity(GO:0004655) |
| 0.0 | 0.1 | GO:0034603 | pyruvate dehydrogenase activity(GO:0004738) pyruvate dehydrogenase [NAD(P)+] activity(GO:0034603) pyruvate dehydrogenase (NAD+) activity(GO:0034604) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.0 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.2 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.2 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |