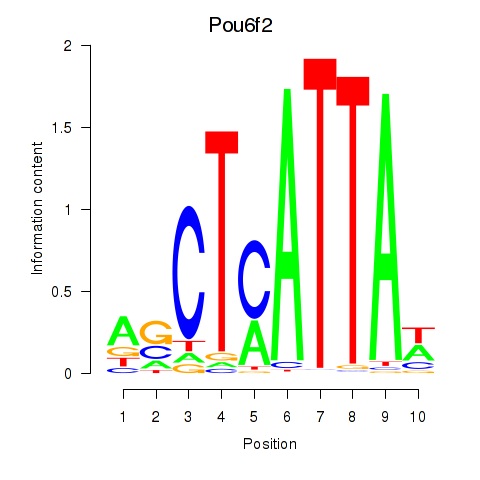
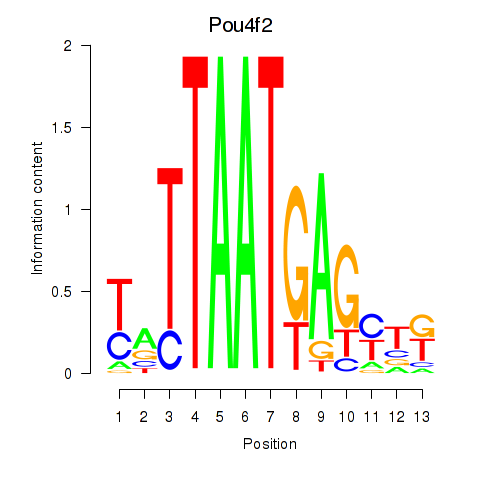
Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Pou6f2_Pou4f2
Z-value: 0.29
Transcription factors associated with Pou6f2_Pou4f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou6f2
|
ENSRNOG00000013237 | POU domain, class 6, transcription factor 2 |
|
Pou4f2
|
ENSRNOG00000012167 | POU class 4 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou6f2 | rn6_v1_chr17_+_49322205_49322205 | -0.50 | 3.9e-01 | Click! |
| Pou4f2 | rn6_v1_chr19_+_33160180_33160180 | -0.20 | 7.5e-01 | Click! |
Activity profile of Pou6f2_Pou4f2 motif
Sorted Z-values of Pou6f2_Pou4f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_2174131 | 0.38 |
ENSRNOT00000001313
|
Pcp2
|
Purkinje cell protein 2 |
| chr3_-_66417741 | 0.15 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr2_+_122877286 | 0.15 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr13_+_71107465 | 0.13 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr12_-_17186679 | 0.13 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr13_+_90723092 | 0.11 |
ENSRNOT00000010146
|
Kcnj10
|
ATP-sensitive inward rectifier potassium channel 10 |
| chr2_+_145174876 | 0.11 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr19_-_58735173 | 0.08 |
ENSRNOT00000030077
|
Pcnx2
|
pecanex homolog 2 (Drosophila) |
| chr12_-_45801842 | 0.08 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr1_+_40879747 | 0.07 |
ENSRNOT00000083702
|
Akap12
|
A-kinase anchoring protein 12 |
| chr10_+_55927223 | 0.06 |
ENSRNOT00000011523
|
Kcnab3
|
potassium voltage-gated channel subfamily A regulatory beta subunit 3 |
| chr2_-_32518643 | 0.06 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_207930796 | 0.06 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr5_+_139790395 | 0.06 |
ENSRNOT00000015033
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr16_+_23668595 | 0.05 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr5_-_12526962 | 0.05 |
ENSRNOT00000092104
|
St18
|
suppression of tumorigenicity 18 |
| chr9_+_71915421 | 0.05 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr1_+_190666149 | 0.05 |
ENSRNOT00000089361
|
LOC102547219
|
uncharacterized LOC102547219 |
| chr4_+_68656928 | 0.04 |
ENSRNOT00000016232
|
Tas2r108
|
taste receptor, type 2, member 108 |
| chr2_+_2729829 | 0.04 |
ENSRNOT00000017068
|
Spata9
|
spermatogenesis associated 9 |
| chr6_+_73553210 | 0.04 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr9_+_73378057 | 0.04 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr6_-_44363915 | 0.04 |
ENSRNOT00000085925
|
Id2
|
inhibitor of DNA binding 2, HLH protein |
| chr6_-_67084234 | 0.04 |
ENSRNOT00000050372
|
Nova1
|
NOVA alternative splicing regulator 1 |
| chr15_-_109394905 | 0.04 |
ENSRNOT00000019602
ENSRNOT00000078424 |
Tmtc4
|
transmembrane and tetratricopeptide repeat containing 4 |
| chr3_+_48106099 | 0.04 |
ENSRNOT00000007218
|
Slc4a10
|
solute carrier family 4 member 10 |
| chr2_-_187786700 | 0.04 |
ENSRNOT00000092257
ENSRNOT00000092612 ENSRNOT00000068360 |
Slc25a44
|
solute carrier family 25, member 44 |
| chr18_+_30880020 | 0.04 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr6_+_146784915 | 0.03 |
ENSRNOT00000008362
|
Sp8
|
Sp8 transcription factor |
| chr18_-_31071371 | 0.03 |
ENSRNOT00000060432
|
Diaph1
|
diaphanous-related formin 1 |
| chr1_+_141767940 | 0.03 |
ENSRNOT00000064034
|
Zfp710
|
zinc finger protein 710 |
| chr12_+_10577068 | 0.03 |
ENSRNOT00000001282
|
Rnf6
|
ring finger protein 6 |
| chr8_+_76754492 | 0.03 |
ENSRNOT00000085727
|
Myo1e
|
myosin IE |
| chr10_-_87286387 | 0.02 |
ENSRNOT00000044206
|
Krt28
|
keratin 28 |
| chr1_-_87825333 | 0.02 |
ENSRNOT00000072689
|
Zfp74
|
zinc finger protein 74 |
| chr3_-_80933283 | 0.02 |
ENSRNOT00000007771
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr8_+_103337472 | 0.02 |
ENSRNOT00000049729
|
Paqr9
|
progestin and adipoQ receptor family member 9 |
| chr18_-_49937522 | 0.02 |
ENSRNOT00000033254
|
Zfp608
|
zinc finger protein 608 |
| chrX_+_86126157 | 0.02 |
ENSRNOT00000006992
|
Klhl4
|
kelch-like family member 4 |
| chr20_+_45458558 | 0.02 |
ENSRNOT00000000713
|
Cdk19
|
cyclin-dependent kinase 19 |
| chr1_+_157920786 | 0.02 |
ENSRNOT00000014284
|
Fam181b
|
family with sequence similarity 181, member B |
| chr3_-_37803112 | 0.02 |
ENSRNOT00000059461
|
Neb
|
nebulin |
| chr8_-_132753145 | 0.02 |
ENSRNOT00000007467
ENSRNOT00000008172 |
RGD1566368
|
similar to Solute carrier family 6 (neurotransmitter transporter), member 20 |
| chrX_-_157200981 | 0.02 |
ENSRNOT00000087364
|
AABR07071935.1
|
|
| chr13_-_60567882 | 0.02 |
ENSRNOT00000004701
|
Trove2
|
TROVE domain family, member 2 |
| chr1_+_217345154 | 0.02 |
ENSRNOT00000092516
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr5_-_109651730 | 0.01 |
ENSRNOT00000093032
|
Elavl2
|
ELAV like RNA binding protein 2 |
| chr4_+_120672152 | 0.01 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr1_-_72311856 | 0.01 |
ENSRNOT00000021286
|
Epn1
|
Epsin 1 |
| chr3_-_57607683 | 0.01 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr18_-_26656879 | 0.01 |
ENSRNOT00000086729
|
Epb41l4a
|
erythrocyte membrane protein band 4.1 like 4A |
| chr8_-_39551700 | 0.01 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr10_-_29450644 | 0.01 |
ENSRNOT00000087937
|
Adra1b
|
adrenoceptor alpha 1B |
| chr4_+_181315444 | 0.01 |
ENSRNOT00000044147
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr15_+_4064706 | 0.01 |
ENSRNOT00000011956
|
Synpo2l
|
synaptopodin 2-like |
| chr5_-_58198782 | 0.01 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr11_-_86303453 | 0.01 |
ENSRNOT00000071453
|
LOC498122
|
similar to CG15908-PA |
| chr19_-_50220455 | 0.01 |
ENSRNOT00000079760
|
Sdr42e1
|
short chain dehydrogenase/reductase family 42E, member 1 |
| chr1_-_134871167 | 0.01 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr1_-_131460473 | 0.01 |
ENSRNOT00000084336
|
Nr2f2
|
nuclear receptor subfamily 2, group F, member 2 |
| chr2_-_205212681 | 0.01 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr5_-_7941822 | 0.01 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr7_+_123260607 | 0.01 |
ENSRNOT00000066849
|
Xrcc6
|
X-ray repair cross complementing 6 |
| chr11_-_62067655 | 0.01 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr6_+_8284878 | 0.00 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr7_+_41475163 | 0.00 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr9_-_73799427 | 0.00 |
ENSRNOT00000067589
|
Kansl1l
|
KAT8 regulatory NSL complex subunit 1-like |
| chr19_+_6046665 | 0.00 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr10_-_87484935 | 0.00 |
ENSRNOT00000016752
|
Krtap3-1
|
keratin associated protein 3-1 |
| chr1_+_261291870 | 0.00 |
ENSRNOT00000049914
|
Hoga1
|
4-hydroxy-2-oxoglutarate aldolase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou6f2_Pou4f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0016056 | rhodopsin mediated signaling pathway(GO:0016056) |
| 0.0 | 0.1 | GO:0051935 | amino acid neurotransmitter reuptake(GO:0051933) glutamate reuptake(GO:0051935) |
| 0.0 | 0.1 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |