Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
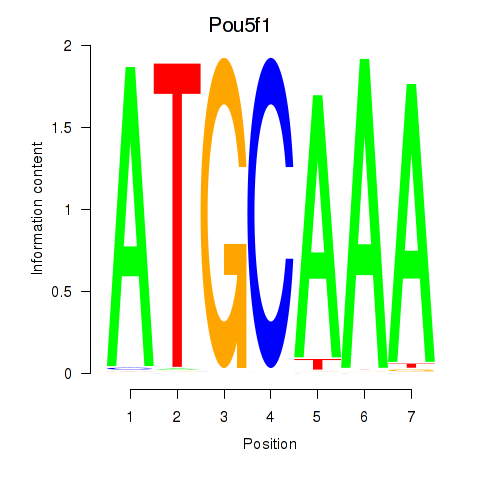
Results for Pou5f1
Z-value: 0.46
Transcription factors associated with Pou5f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou5f1
|
ENSRNOG00000046487 | POU class 5 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou5f1 | rn6_v1_chr20_-_3751994_3751994 | 0.30 | 6.3e-01 | Click! |
Activity profile of Pou5f1 motif
Sorted Z-values of Pou5f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr20_-_22459025 | 0.36 |
ENSRNOT00000000792
|
Egr2
|
early growth response 2 |
| chr10_+_77537340 | 0.31 |
ENSRNOT00000003297
|
Tmem100
|
transmembrane protein 100 |
| chr1_+_81372650 | 0.21 |
ENSRNOT00000088829
|
Zfp428
|
zinc finger protein 428 |
| chr17_+_49417067 | 0.21 |
ENSRNOT00000090024
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr17_+_43632397 | 0.19 |
ENSRNOT00000013790
|
Hist1h2ah
|
histone cluster 1, H2ah |
| chr2_-_198360678 | 0.18 |
ENSRNOT00000051917
|
Hist2h2ac
|
histone cluster 2 H2A family member c |
| chr17_-_44527801 | 0.16 |
ENSRNOT00000089643
|
Hist1h2bk
|
histone cluster 1 H2B family member k |
| chr1_-_88111293 | 0.15 |
ENSRNOT00000077195
|
Spred3
|
sprouty-related, EVH1 domain containing 3 |
| chr2_-_139528162 | 0.14 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
| chr18_-_12640716 | 0.14 |
ENSRNOT00000020697
|
Klhl14
|
kelch-like family member 14 |
| chr1_-_218100272 | 0.13 |
ENSRNOT00000028411
ENSRNOT00000088588 |
Ccnd1
|
cyclin D1 |
| chr8_-_117932518 | 0.12 |
ENSRNOT00000028130
|
Camp
|
cathelicidin antimicrobial peptide |
| chr15_-_62200837 | 0.12 |
ENSRNOT00000017599
|
Pcdh8
|
protocadherin 8 |
| chr7_+_3133506 | 0.12 |
ENSRNOT00000049972
|
Pmel
|
premelanosome protein |
| chr3_+_79918969 | 0.11 |
ENSRNOT00000016306
|
Spi1
|
Spi-1 proto-oncogene |
| chr1_-_260254600 | 0.11 |
ENSRNOT00000019014
|
Blnk
|
B-cell linker |
| chr1_+_64101018 | 0.11 |
ENSRNOT00000078640
|
Mboat7
|
membrane bound O-acyltransferase domain containing 7 |
| chr10_+_55687050 | 0.10 |
ENSRNOT00000057136
|
Per1
|
period circadian clock 1 |
| chrX_+_80213332 | 0.10 |
ENSRNOT00000042827
|
Sh3bgrl
|
SH3 domain binding glutamate-rich protein like |
| chr7_+_28412198 | 0.10 |
ENSRNOT00000081822
ENSRNOT00000038780 ENSRNOT00000005995 |
Igf1
|
insulin-like growth factor 1 |
| chr16_-_20486707 | 0.09 |
ENSRNOT00000026470
|
Jund
|
JunD proto-oncogene, AP-1 transcription factor subunit |
| chr10_-_50402616 | 0.09 |
ENSRNOT00000004546
|
Hs3st3b1
|
heparan sulfate-glucosamine 3-sulfotransferase 3B1 |
| chr1_-_57327379 | 0.09 |
ENSRNOT00000080429
|
Dll1
|
delta like canonical Notch ligand 1 |
| chr1_+_72545596 | 0.08 |
ENSRNOT00000022698
|
Shisa7
|
shisa family member 7 |
| chr5_+_145257714 | 0.08 |
ENSRNOT00000019214
|
Dlgap3
|
DLG associated protein 3 |
| chr13_-_60849094 | 0.08 |
ENSRNOT00000005156
|
Rgs2
|
regulator of G-protein signaling 2 |
| chr2_+_182723854 | 0.08 |
ENSRNOT00000012658
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr5_-_79874671 | 0.08 |
ENSRNOT00000084563
|
Tnc
|
tenascin C |
| chr4_+_102351036 | 0.08 |
ENSRNOT00000079277
|
AABR07060994.1
|
|
| chr7_+_12619774 | 0.08 |
ENSRNOT00000015257
|
Med16
|
mediator complex subunit 16 |
| chr2_+_222214885 | 0.08 |
ENSRNOT00000055723
|
Dpyd
|
dihydropyrimidine dehydrogenase |
| chr10_+_98706960 | 0.08 |
ENSRNOT00000006217
|
Map2k6
|
mitogen-activated protein kinase kinase 6 |
| chr20_-_5155293 | 0.08 |
ENSRNOT00000092322
|
Prrc2a
|
proline-rich coiled-coil 2A |
| chr17_+_15749978 | 0.08 |
ENSRNOT00000067311
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr15_+_12827707 | 0.08 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr10_+_45297937 | 0.07 |
ENSRNOT00000066367
|
Trim17
|
tripartite motif-containing 17 |
| chr8_+_64364741 | 0.07 |
ENSRNOT00000082840
|
Celf6
|
CUGBP, Elav-like family member 6 |
| chr15_-_14737704 | 0.07 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr2_-_250232295 | 0.07 |
ENSRNOT00000082132
|
Lmo4
|
LIM domain only 4 |
| chr18_-_37776453 | 0.07 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr2_-_25235275 | 0.07 |
ENSRNOT00000061580
|
F2rl1
|
F2R like trypsin receptor 1 |
| chr10_-_108196217 | 0.07 |
ENSRNOT00000075440
|
Cbx4
|
chromobox 4 |
| chr1_-_88690534 | 0.06 |
ENSRNOT00000072570
|
Ovol3
|
ovo-like zinc finger 3 |
| chr5_+_64294321 | 0.06 |
ENSRNOT00000083796
|
Msantd3
|
Myb/SANT DNA binding domain containing 3 |
| chr10_-_45297385 | 0.06 |
ENSRNOT00000041187
|
Hist3h2bb
|
histone cluster 3 H2B family member b |
| chr13_-_89874008 | 0.06 |
ENSRNOT00000051368
|
Ptma
|
prothymosin alpha |
| chr10_-_86004096 | 0.06 |
ENSRNOT00000091978
ENSRNOT00000066855 |
Stac2
|
SH3 and cysteine rich domain 2 |
| chr18_-_37245809 | 0.06 |
ENSRNOT00000079585
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr7_+_97559841 | 0.06 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr18_-_36893797 | 0.06 |
ENSRNOT00000043544
|
Gpr151
|
G protein-coupled receptor 151 |
| chr2_+_32820322 | 0.06 |
ENSRNOT00000013768
|
Cd180
|
CD180 molecule |
| chr18_+_65155685 | 0.06 |
ENSRNOT00000081797
|
Tcf4
|
transcription factor 4 |
| chr2_-_89310946 | 0.05 |
ENSRNOT00000015195
|
Ralyl
|
RALY RNA binding protein-like |
| chr3_+_2396143 | 0.05 |
ENSRNOT00000012388
|
Nrarp
|
Notch-regulated ankyrin repeat protein |
| chr1_+_87938042 | 0.05 |
ENSRNOT00000027837
|
Map4k1
|
mitogen activated protein kinase kinase kinase kinase 1 |
| chr1_+_151439409 | 0.05 |
ENSRNOT00000022060
ENSRNOT00000050639 |
Grm5
|
glutamate metabotropic receptor 5 |
| chr5_+_16526058 | 0.05 |
ENSRNOT00000011130
|
Lyn
|
LYN proto-oncogene, Src family tyrosine kinase |
| chr7_-_104541392 | 0.05 |
ENSRNOT00000078116
|
Fam49b
|
family with sequence similarity 49, member B |
| chr18_+_44737154 | 0.05 |
ENSRNOT00000021972
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr1_+_91038886 | 0.05 |
ENSRNOT00000041301
|
Ovol3
|
ovo-like zinc finger 3 |
| chrX_-_30831483 | 0.05 |
ENSRNOT00000004443
|
Glra2
|
glycine receptor, alpha 2 |
| chr4_-_55011415 | 0.05 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr5_+_59228199 | 0.05 |
ENSRNOT00000060282
|
Hrct1
|
histidine rich carboxyl terminus 1 |
| chr4_+_142453013 | 0.05 |
ENSRNOT00000056573
|
AABR07061755.1
|
|
| chr20_+_5646097 | 0.05 |
ENSRNOT00000090925
|
Itpr3
|
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr9_+_16737642 | 0.05 |
ENSRNOT00000061430
|
Srf
|
serum response factor |
| chr3_+_98297554 | 0.05 |
ENSRNOT00000006524
|
Kcna4
|
potassium voltage-gated channel subfamily A member 4 |
| chr14_+_104191517 | 0.04 |
ENSRNOT00000006573
|
Spred2
|
sprouty-related, EVH1 domain containing 2 |
| chr7_+_123381077 | 0.04 |
ENSRNOT00000082603
ENSRNOT00000056041 |
Srebf2
|
sterol regulatory element binding transcription factor 2 |
| chr14_-_83741969 | 0.04 |
ENSRNOT00000026293
|
Inpp5j
|
inositol polyphosphate-5-phosphatase J |
| chr16_+_46731403 | 0.04 |
ENSRNOT00000017624
|
Tenm3
|
teneurin transmembrane protein 3 |
| chr10_-_94500591 | 0.04 |
ENSRNOT00000015976
|
Cd79b
|
CD79b molecule |
| chr9_-_54457753 | 0.04 |
ENSRNOT00000020032
|
Stat1
|
signal transducer and activator of transcription 1 |
| chr2_+_121165137 | 0.04 |
ENSRNOT00000016236
|
Sox2
|
SRY box 2 |
| chr3_-_3951025 | 0.04 |
ENSRNOT00000026212
|
Notch1
|
notch 1 |
| chr3_+_93495106 | 0.04 |
ENSRNOT00000029922
ENSRNOT00000085760 |
Abtb2
|
ankyrin repeat and BTB domain containing 2 |
| chr3_+_150910398 | 0.04 |
ENSRNOT00000055310
|
Tp53inp2
|
tumor protein p53 inducible nuclear protein 2 |
| chr9_-_23493081 | 0.04 |
ENSRNOT00000072144
|
Rhag
|
Rh-associated glycoprotein |
| chr3_-_152179193 | 0.03 |
ENSRNOT00000026700
|
Rbm12
|
RNA binding motif protein 12 |
| chr5_+_154489590 | 0.03 |
ENSRNOT00000035788
|
Id3
|
inhibitor of DNA binding 3, HLH protein |
| chr4_-_157266018 | 0.03 |
ENSRNOT00000019570
|
LOC100911713
|
protein C10-like |
| chr8_+_93491761 | 0.03 |
ENSRNOT00000014326
|
Tpbg
|
trophoblast glycoprotein |
| chr4_+_120672152 | 0.03 |
ENSRNOT00000077231
|
Mgll
|
monoglyceride lipase |
| chr16_-_49820235 | 0.03 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr2_-_179704629 | 0.03 |
ENSRNOT00000083361
ENSRNOT00000077941 |
Gria2
|
glutamate ionotropic receptor AMPA type subunit 2 |
| chr9_+_93545396 | 0.03 |
ENSRNOT00000025093
|
LOC100359583
|
hypothetical protein LOC100359583 |
| chr2_+_243502073 | 0.03 |
ENSRNOT00000015870
|
Adh7
|
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr4_-_85915099 | 0.03 |
ENSRNOT00000016182
|
Neurod6
|
neuronal differentiation 6 |
| chr4_-_119327822 | 0.03 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr20_+_7788084 | 0.03 |
ENSRNOT00000000597
|
Def6
|
DEF6 guanine nucleotide exchange factor |
| chr15_-_43542939 | 0.03 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr3_-_2411544 | 0.03 |
ENSRNOT00000012406
|
Tor4a
|
torsin family 4, member A |
| chr11_+_3119885 | 0.03 |
ENSRNOT00000042316
|
Vgll3
|
vestigial-like family member 3 |
| chr2_+_198418691 | 0.03 |
ENSRNOT00000089409
|
Hist1h2bk
|
histone cluster 1, H2bk |
| chr2_-_231877307 | 0.03 |
ENSRNOT00000073860
|
Larp7
|
La ribonucleoprotein domain family, member 7 |
| chr8_+_93492122 | 0.03 |
ENSRNOT00000089491
|
Tpbg
|
trophoblast glycoprotein |
| chr15_+_58016238 | 0.03 |
ENSRNOT00000075786
|
Kctd4
|
potassium channel tetramerization domain containing 4 |
| chr10_-_87335823 | 0.03 |
ENSRNOT00000079297
|
Krt12
|
keratin 12 |
| chr18_+_53915807 | 0.03 |
ENSRNOT00000026543
|
Adamts19
|
ADAM metallopeptidase with thrombospondin type 1 motif, 19 |
| chr2_+_55835151 | 0.03 |
ENSRNOT00000018634
|
Fyb
|
FYN binding protein |
| chr2_+_198360998 | 0.03 |
ENSRNOT00000046129
|
Hist2h2be
|
histone cluster 2, H2be |
| chr8_+_54993859 | 0.03 |
ENSRNOT00000013093
|
Il18
|
interleukin 18 |
| chr2_+_174542667 | 0.03 |
ENSRNOT00000076793
|
Fstl5
|
follistatin-like 5 |
| chr10_-_86690815 | 0.03 |
ENSRNOT00000012537
|
Nr1d1
|
nuclear receptor subfamily 1, group D, member 1 |
| chr5_+_140870140 | 0.03 |
ENSRNOT00000074347
|
Hpcal4
|
hippocalcin-like 4 |
| chr1_+_225140863 | 0.02 |
ENSRNOT00000027141
|
Mta2
|
metastasis associated 1 family, member 2 |
| chr1_+_144070754 | 0.02 |
ENSRNOT00000079989
|
Sh3gl3
|
SH3 domain-containing GRB2-like 3 |
| chr4_-_183424449 | 0.02 |
ENSRNOT00000071930
|
Fam60a
|
family with sequence similarity 60, member A |
| chr17_+_44520537 | 0.02 |
ENSRNOT00000077985
|
hist1h2ail2
|
histone cluster 1, H2ai-like2 |
| chrX_-_15467875 | 0.02 |
ENSRNOT00000011207
|
Pim2
|
Pim-2 proto-oncogene, serine/threonine kinase |
| chr1_-_87937516 | 0.02 |
ENSRNOT00000087522
|
Eif3k
|
eukaryotic translation initiation factor 3, subunit K |
| chr4_-_125808281 | 0.02 |
ENSRNOT00000037848
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr20_+_25990656 | 0.02 |
ENSRNOT00000081254
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr1_-_255376833 | 0.02 |
ENSRNOT00000024941
|
Ppp1r3c
|
protein phosphatase 1, regulatory subunit 3C |
| chr6_+_124217241 | 0.02 |
ENSRNOT00000064679
|
Calm3
|
calmodulin 3 |
| chr12_+_38160464 | 0.02 |
ENSRNOT00000032249
|
Hcar2
|
hydroxycarboxylic acid receptor 2 |
| chrX_+_159505344 | 0.02 |
ENSRNOT00000001164
|
Brs3
|
bombesin receptor subtype 3 |
| chrX_-_107442878 | 0.02 |
ENSRNOT00000052302
|
Glra4
|
glycine receptor, alpha 4 |
| chr6_-_55524436 | 0.02 |
ENSRNOT00000006752
|
Tspan13
|
tetraspanin 13 |
| chr3_-_17081510 | 0.02 |
ENSRNOT00000063862
|
AABR07051562.1
|
|
| chr10_-_37311625 | 0.02 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr2_+_18354542 | 0.02 |
ENSRNOT00000042958
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr17_+_52938980 | 0.02 |
ENSRNOT00000081052
|
Gabpb1l
|
GA binding protein transcription factor, beta subunit 1-like |
| chr7_-_60416925 | 0.02 |
ENSRNOT00000008193
|
Cpsf6
|
cleavage and polyadenylation specific factor 6 |
| chr2_+_116970344 | 0.02 |
ENSRNOT00000039603
|
Egfem1
|
EGF-like and EMI domain containing 1 |
| chr1_+_80141630 | 0.02 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr16_-_20939545 | 0.02 |
ENSRNOT00000027457
|
Sugp2
|
SURP and G patch domain containing 2 |
| chr3_+_116899878 | 0.02 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr8_-_59239954 | 0.02 |
ENSRNOT00000016104
|
Acsbg1
|
acyl-CoA synthetase bubblegum family member 1 |
| chr14_-_45165207 | 0.02 |
ENSRNOT00000002960
|
Klf3
|
Kruppel like factor 3 |
| chr17_-_43689311 | 0.02 |
ENSRNOT00000028779
|
Hist1h2bcl1
|
histone cluster 1, H2bc-like 1 |
| chr18_+_61261418 | 0.02 |
ENSRNOT00000064250
|
Zfp532
|
zinc finger protein 532 |
| chr8_-_111850393 | 0.02 |
ENSRNOT00000044956
|
Cdv3
|
CDV3 homolog |
| chr19_-_25062934 | 0.01 |
ENSRNOT00000060117
|
Gm10644
|
predicted gene 10644 |
| chr20_+_25990304 | 0.01 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr7_-_11443357 | 0.01 |
ENSRNOT00000082126
|
Zbtb7a
|
zinc finger and BTB domain containing 7a |
| chr2_+_18392142 | 0.01 |
ENSRNOT00000043196
|
Hapln1
|
hyaluronan and proteoglycan link protein 1 |
| chr1_+_268189277 | 0.01 |
ENSRNOT00000065001
|
Sorcs3
|
sortilin-related VPS10 domain containing receptor 3 |
| chr12_+_37544918 | 0.01 |
ENSRNOT00000001403
|
Rilpl2
|
Rab interacting lysosomal protein-like 2 |
| chr19_+_50848736 | 0.01 |
ENSRNOT00000077053
|
Cdh13
|
cadherin 13 |
| chr13_-_61003744 | 0.01 |
ENSRNOT00000005163
|
Rgs13
|
regulator of G-protein signaling 13 |
| chr1_-_251379593 | 0.01 |
ENSRNOT00000014684
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr4_-_148048726 | 0.01 |
ENSRNOT00000042418
ENSRNOT00000056393 |
Tmcc1
|
transmembrane and coiled-coil domain family 1 |
| chr8_-_78397123 | 0.01 |
ENSRNOT00000087270
ENSRNOT00000084925 |
Tcf12
|
transcription factor 12 |
| chr12_-_38995570 | 0.01 |
ENSRNOT00000001806
|
Orai1
|
ORAI calcium release-activated calcium modulator 1 |
| chr9_+_73529612 | 0.01 |
ENSRNOT00000032430
|
Unc80
|
unc-80 homolog, NALCN activator |
| chr5_+_2353468 | 0.01 |
ENSRNOT00000090346
|
Stau2
|
staufen double-stranded RNA binding protein 2 |
| chr6_-_37122023 | 0.01 |
ENSRNOT00000007609
|
Vsnl1
|
visinin-like 1 |
| chr8_-_112648880 | 0.01 |
ENSRNOT00000015265
|
Ackr4
|
atypical chemokine receptor 4 |
| chr4_+_147333056 | 0.01 |
ENSRNOT00000012137
|
Pparg
|
peroxisome proliferator-activated receptor gamma |
| chr17_-_67904674 | 0.01 |
ENSRNOT00000078532
|
Klf6
|
Kruppel-like factor 6 |
| chr2_-_173563273 | 0.01 |
ENSRNOT00000081423
|
Zbbx
|
zinc finger, B-box domain containing |
| chr2_-_250805445 | 0.01 |
ENSRNOT00000055362
|
Clca4l
|
chloride channel calcium activated 4-like |
| chr2_-_184309664 | 0.01 |
ENSRNOT00000052051
|
Fbxw7
|
F-box and WD repeat domain containing 7 |
| chr10_-_8498422 | 0.01 |
ENSRNOT00000082332
|
Rbfox1
|
RNA binding protein, fox-1 homolog 1 |
| chr10_+_86657285 | 0.01 |
ENSRNOT00000087346
|
Thra
|
thyroid hormone receptor alpha |
| chr6_-_61405195 | 0.01 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr17_+_72160735 | 0.01 |
ENSRNOT00000038817
|
Itih2
|
inter-alpha-trypsin inhibitor heavy chain 2 |
| chr4_-_18396035 | 0.00 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr1_+_252894663 | 0.00 |
ENSRNOT00000054757
|
Ifit2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr2_-_250778269 | 0.00 |
ENSRNOT00000085670
ENSRNOT00000083535 ENSRNOT00000017824 |
Clca5
|
chloride channel accessory 5 |
| chr2_-_149088787 | 0.00 |
ENSRNOT00000064833
|
Clrn1
|
clarin 1 |
| chr8_-_71337746 | 0.00 |
ENSRNOT00000021554
|
Zfp609
|
zinc finger protein 609 |
| chr10_-_74119009 | 0.00 |
ENSRNOT00000085712
ENSRNOT00000006926 |
Dhx40
|
DEAH-box helicase 40 |
| chr13_+_57131395 | 0.00 |
ENSRNOT00000017884
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr10_+_89285855 | 0.00 |
ENSRNOT00000028033
|
G6pc
|
glucose-6-phosphatase, catalytic subunit |
| chr1_+_277068761 | 0.00 |
ENSRNOT00000044183
ENSRNOT00000022382 |
Habp2
|
hyaluronan binding protein 2 |
| chr6_-_138632159 | 0.00 |
ENSRNOT00000082921
ENSRNOT00000040702 |
Ighm
|
immunoglobulin heavy constant mu |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou5f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0021593 | rhombomere morphogenesis(GO:0021593) |
| 0.1 | 0.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.0 | 0.1 | GO:0048023 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.1 | GO:0045347 | negative regulation of MHC class II biosynthetic process(GO:0045347) |
| 0.0 | 0.1 | GO:2000041 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0034136 | negative regulation of toll-like receptor 2 signaling pathway(GO:0034136) |
| 0.0 | 0.1 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.1 | GO:0006214 | thymidine catabolic process(GO:0006214) pyrimidine deoxyribonucleoside catabolic process(GO:0046127) |
| 0.0 | 0.1 | GO:0001834 | trophectodermal cell proliferation(GO:0001834) |
| 0.0 | 0.1 | GO:0043311 | negative regulation of toll-like receptor 3 signaling pathway(GO:0034140) regulation of eosinophil degranulation(GO:0043309) positive regulation of eosinophil degranulation(GO:0043311) regulation of neutrophil mediated cytotoxicity(GO:0070948) regulation of neutrophil mediated killing of symbiont cell(GO:0070949) positive regulation of renin secretion into blood stream(GO:1900135) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.0 | GO:0090370 | negative regulation of cholesterol efflux(GO:0090370) |
| 0.0 | 0.1 | GO:0000320 | re-entry into mitotic cell cycle(GO:0000320) |
| 0.0 | 0.3 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 0.1 | GO:0071799 | bud outgrowth involved in lung branching(GO:0060447) response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.1 | GO:0099552 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0035709 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.0 | GO:0046016 | positive regulation of transcription by glucose(GO:0046016) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.1 | GO:0071877 | regulation of adrenergic receptor signaling pathway(GO:0071877) |
| 0.0 | 0.0 | GO:0034240 | negative regulation of macrophage fusion(GO:0034240) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032585 | multivesicular body membrane(GO:0032585) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.0 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.0 | GO:0019815 | B cell receptor complex(GO:0019815) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.0 | 0.1 | GO:0004159 | dihydrouracil dehydrogenase (NAD+) activity(GO:0004159) dihydropyrimidine dehydrogenase (NADP+) activity(GO:0017113) |
| 0.0 | 0.1 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.0 | 0.0 | GO:0022852 | glycine-gated chloride ion channel activity(GO:0022852) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0052743 | phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) inositol tetrakisphosphate phosphatase activity(GO:0052743) |
| 0.0 | 0.1 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.0 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.0 | 0.1 | GO:0061133 | endopeptidase activator activity(GO:0061133) |
| 0.0 | 0.0 | GO:0004971 | AMPA glutamate receptor activity(GO:0004971) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | ST PAC1 RECEPTOR PATHWAY | PAC1 Receptor Pathway |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME PACKAGING OF TELOMERE ENDS | Genes involved in Packaging Of Telomere Ends |