Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
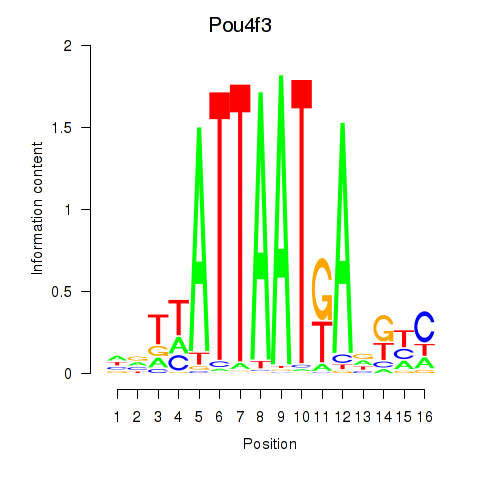
Results for Pou4f3
Z-value: 0.32
Transcription factors associated with Pou4f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou4f3
|
ENSRNOG00000018842 | POU class 4 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou4f3 | rn6_v1_chr18_+_36713869_36713869 | 0.08 | 8.9e-01 | Click! |
Activity profile of Pou4f3 motif
Sorted Z-values of Pou4f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_190370499 | 0.20 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr10_+_61685645 | 0.16 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr2_+_127525285 | 0.13 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr4_-_18396035 | 0.10 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr17_-_79085076 | 0.09 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr2_+_122877286 | 0.09 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr19_+_24800072 | 0.08 |
ENSRNOT00000005470
|
Ptger1
|
prostaglandin E receptor 1 |
| chr9_+_73378057 | 0.08 |
ENSRNOT00000043627
ENSRNOT00000045766 ENSRNOT00000092445 ENSRNOT00000037974 |
Map2
|
microtubule-associated protein 2 |
| chr12_+_10577068 | 0.07 |
ENSRNOT00000001282
|
Rnf6
|
ring finger protein 6 |
| chr18_+_30895831 | 0.07 |
ENSRNOT00000026985
|
Pcdhga10
|
protocadherin gamma subfamily A, 10 |
| chr18_+_14757679 | 0.07 |
ENSRNOT00000071364
|
Mapre2
|
microtubule-associated protein, RP/EB family, member 2 |
| chr4_-_66955732 | 0.06 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr5_-_161875362 | 0.06 |
ENSRNOT00000078237
|
Prdm2
|
PR/SET domain 2 |
| chr19_+_14835822 | 0.05 |
ENSRNOT00000072804
|
1700007B14Rik
|
RIKEN cDNA 1700007B14 gene |
| chr5_-_12199283 | 0.05 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr7_+_120923274 | 0.05 |
ENSRNOT00000049247
|
Gtpbp1
|
GTP binding protein 1 |
| chr1_-_166912524 | 0.05 |
ENSRNOT00000092952
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr17_+_9596957 | 0.04 |
ENSRNOT00000017349
|
Fam193b
|
family with sequence similarity 193, member B |
| chr18_+_30869628 | 0.04 |
ENSRNOT00000060470
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr18_+_30880020 | 0.04 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr12_-_35979193 | 0.04 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr17_-_4454701 | 0.04 |
ENSRNOT00000080750
ENSRNOT00000066950 |
Dapk1
|
death associated protein kinase 1 |
| chr12_-_46493203 | 0.04 |
ENSRNOT00000057036
|
Cit
|
citron rho-interacting serine/threonine kinase |
| chr10_+_29606748 | 0.03 |
ENSRNOT00000080720
|
AABR07029467.2
|
|
| chr7_+_71157664 | 0.03 |
ENSRNOT00000005919
|
Sdr9c7
|
short chain dehydrogenase/reductase family 9C, member 7 |
| chr8_-_126390801 | 0.03 |
ENSRNOT00000089732
|
AABR07071661.1
|
|
| chr14_-_19072677 | 0.03 |
ENSRNOT00000060548
|
LOC360919
|
similar to alpha-fetoprotein |
| chr16_-_20562399 | 0.03 |
ENSRNOT00000031692
|
Lrrc25
|
leucine rich repeat containing 25 |
| chr2_-_198706428 | 0.02 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr18_-_43945273 | 0.02 |
ENSRNOT00000088900
|
Dtwd2
|
DTW domain containing 2 |
| chr7_+_117456039 | 0.02 |
ENSRNOT00000051754
|
Mroh1
|
maestro heat-like repeat family member 1 |
| chr8_+_118206698 | 0.02 |
ENSRNOT00000082607
ENSRNOT00000064140 |
Smarcc1
|
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 1 |
| chrX_+_65040934 | 0.02 |
ENSRNOT00000044006
|
Zc3h12b
|
zinc finger CCCH-type containing 12B |
| chr6_+_42947334 | 0.02 |
ENSRNOT00000041376
|
RGD1563157
|
similar to 60S ribosomal protein L35 |
| chr8_-_57830504 | 0.02 |
ENSRNOT00000017683
|
Ddx10
|
DEAD-box helicase 10 |
| chr4_-_180505916 | 0.02 |
ENSRNOT00000086465
|
AABR07062512.1
|
|
| chr3_+_55910177 | 0.02 |
ENSRNOT00000009969
|
Klhl41
|
kelch-like family member 41 |
| chr7_-_140291620 | 0.02 |
ENSRNOT00000088323
|
Adcy6
|
adenylate cyclase 6 |
| chr12_+_47218969 | 0.02 |
ENSRNOT00000081343
ENSRNOT00000038395 |
Unc119b
|
unc-119 lipid binding chaperone B |
| chr16_+_84465656 | 0.02 |
ENSRNOT00000043188
|
LOC689713
|
LRRGT00175 |
| chr13_-_102857551 | 0.02 |
ENSRNOT00000080309
|
Mark1
|
microtubule affinity regulating kinase 1 |
| chr2_-_235257731 | 0.02 |
ENSRNOT00000078074
|
Gar1
|
GAR1 ribonucleoprotein |
| chr19_+_6046665 | 0.01 |
ENSRNOT00000084126
|
Cdh8
|
cadherin 8 |
| chr3_-_80933283 | 0.01 |
ENSRNOT00000007771
|
Creb3l1
|
cAMP responsive element binding protein 3-like 1 |
| chr10_+_103206014 | 0.01 |
ENSRNOT00000004081
|
Ttyh2
|
tweety family member 2 |
| chrX_-_76925195 | 0.01 |
ENSRNOT00000087977
|
Atrx
|
ATRX, chromatin remodeler |
| chr1_-_164977633 | 0.01 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr18_+_17043903 | 0.01 |
ENSRNOT00000068139
|
Fhod3
|
formin homology 2 domain containing 3 |
| chr6_+_123034304 | 0.01 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
| chr3_-_21904133 | 0.01 |
ENSRNOT00000090576
ENSRNOT00000087611 ENSRNOT00000066377 |
Strbp
|
spermatid perinuclear RNA binding protein |
| chr9_+_52687868 | 0.00 |
ENSRNOT00000083864
|
Wdr75
|
WD repeat domain 75 |
| chr6_-_61405195 | 0.00 |
ENSRNOT00000008655
|
Lrrn3
|
leucine rich repeat neuronal 3 |
| chr10_-_89088993 | 0.00 |
ENSRNOT00000027458
|
Ccr10
|
C-C motif chemokine receptor 10 |
| chr1_+_174702373 | 0.00 |
ENSRNOT00000013733
|
Zfp143
|
zinc finger protein 143 |
| chr17_-_76281660 | 0.00 |
ENSRNOT00000058070
|
Upf2
|
UPF2 regulator of nonsense transcripts homolog (yeast) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou4f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1903375 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) hypothalamus gonadotrophin-releasing hormone neuron differentiation(GO:0021886) hypothalamus gonadotrophin-releasing hormone neuron development(GO:0021888) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.1 | GO:0044314 | protein K27-linked ubiquitination(GO:0044314) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0004957 | prostaglandin E receptor activity(GO:0004957) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |