Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
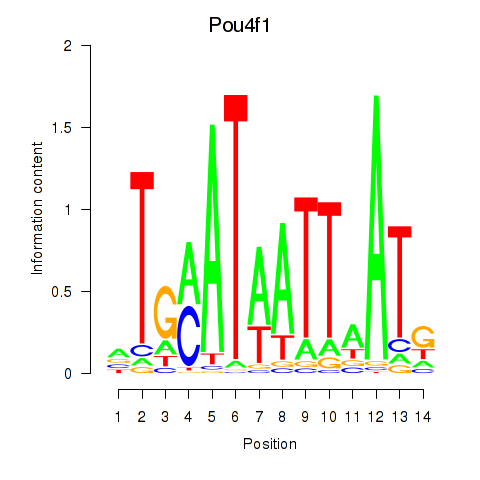
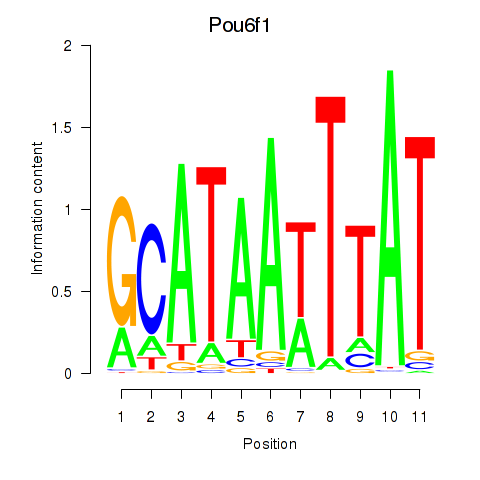
Results for Pou4f1_Pou6f1
Z-value: 0.67
Transcription factors associated with Pou4f1_Pou6f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou4f1
|
ENSRNOG00000060662 | POU class 4 homeobox 1 |
|
Pou6f1
|
ENSRNOG00000004595 | POU class 6 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou4f1 | rn6_v1_chr15_-_88622413_88622413 | -0.75 | 1.5e-01 | Click! |
| Pou6f1 | rn6_v1_chr7_-_142210738_142210738 | 0.20 | 7.4e-01 | Click! |
Activity profile of Pou4f1_Pou6f1 motif
Sorted Z-values of Pou4f1_Pou6f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_41467064 | 0.39 |
ENSRNOT00000073231
|
AABR07008066.2
|
|
| chr14_+_71533063 | 0.28 |
ENSRNOT00000004231
|
Prom1
|
prominin 1 |
| chr14_-_55081551 | 0.24 |
ENSRNOT00000049245
|
Pcdh7
|
protocadherin 7 |
| chr2_+_180012414 | 0.23 |
ENSRNOT00000082273
|
Pdgfc
|
platelet derived growth factor C |
| chr5_+_25253010 | 0.22 |
ENSRNOT00000061333
|
Rbm12b
|
RNA binding motif protein 12B |
| chr5_+_27326762 | 0.22 |
ENSRNOT00000066191
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr8_-_123829749 | 0.21 |
ENSRNOT00000090467
|
AABR07071598.1
|
|
| chr5_+_25725683 | 0.21 |
ENSRNOT00000087602
|
LOC679087
|
similar to swan |
| chr18_-_70924708 | 0.21 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr4_+_22859622 | 0.20 |
ENSRNOT00000073501
ENSRNOT00000068410 |
Adam22
|
ADAM metallopeptidase domain 22 |
| chr2_-_219262901 | 0.19 |
ENSRNOT00000037068
|
Gpr88
|
G-protein coupled receptor 88 |
| chr10_+_14136883 | 0.18 |
ENSRNOT00000082295
|
AC115181.1
|
|
| chr19_+_43163129 | 0.18 |
ENSRNOT00000073721
|
Clec18a
|
C-type lectin domain family 18, member A |
| chr14_-_41786084 | 0.17 |
ENSRNOT00000059439
|
Grxcr1
|
glutaredoxin and cysteine rich domain containing 1 |
| chr7_+_74350479 | 0.16 |
ENSRNOT00000089034
|
AABR07057495.1
|
|
| chr8_-_63291966 | 0.15 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr1_+_167169442 | 0.15 |
ENSRNOT00000048172
|
LOC102549471
|
short transient receptor potential channel 2-like |
| chr17_-_86657473 | 0.15 |
ENSRNOT00000078827
|
AABR07028795.1
|
|
| chr2_-_35104963 | 0.15 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr14_+_71533460 | 0.15 |
ENSRNOT00000084274
|
Prom1
|
prominin 1 |
| chr6_+_27328406 | 0.14 |
ENSRNOT00000091159
ENSRNOT00000044278 |
Otof
|
otoferlin |
| chrX_+_115208863 | 0.14 |
ENSRNOT00000072094
|
AABR07040936.1
|
|
| chrX_-_142164220 | 0.14 |
ENSRNOT00000064780
|
Fgf13
|
fibroblast growth factor 13 |
| chr16_-_81880502 | 0.14 |
ENSRNOT00000079096
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr1_-_280015358 | 0.14 |
ENSRNOT00000024548
|
Hspa12a
|
heat shock protein family A (Hsp70) member 12A |
| chr8_-_93390305 | 0.13 |
ENSRNOT00000056930
|
Ibtk
|
inhibitor of Bruton tyrosine kinase |
| chr6_+_64789940 | 0.13 |
ENSRNOT00000085979
ENSRNOT00000059739 ENSRNOT00000051908 ENSRNOT00000082793 ENSRNOT00000078583 ENSRNOT00000091677 ENSRNOT00000093241 |
Nrcam
|
neuronal cell adhesion molecule |
| chr16_+_39827749 | 0.13 |
ENSRNOT00000074885
|
Wdr17
|
WD repeat domain 17 |
| chr13_-_51784639 | 0.13 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr12_-_19254527 | 0.13 |
ENSRNOT00000089349
|
AABR07035541.3
|
|
| chr1_-_19376301 | 0.13 |
ENSRNOT00000015547
|
Arhgap18
|
Rho GTPase activating protein 18 |
| chr7_-_68549763 | 0.13 |
ENSRNOT00000078014
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr10_+_73868943 | 0.12 |
ENSRNOT00000081012
|
Tubd1
|
tubulin, delta 1 |
| chr15_+_1054937 | 0.12 |
ENSRNOT00000008154
|
AABR07016841.1
|
|
| chr4_-_165456677 | 0.12 |
ENSRNOT00000082207
|
Klra2
|
killer cell lectin-like receptor, subfamily A, member 2 |
| chrX_+_88298266 | 0.11 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr9_+_73334618 | 0.11 |
ENSRNOT00000092717
|
Map2
|
microtubule-associated protein 2 |
| chr7_-_124929025 | 0.11 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr4_-_95970666 | 0.11 |
ENSRNOT00000008826
|
Hpgds
|
hematopoietic prostaglandin D synthase |
| chr3_-_150064438 | 0.11 |
ENSRNOT00000086933
|
E2f1
|
E2F transcription factor 1 |
| chr8_+_107882219 | 0.10 |
ENSRNOT00000019902
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr9_+_2190915 | 0.10 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr1_-_246010594 | 0.10 |
ENSRNOT00000023762
|
Rfx3
|
regulatory factor X3 |
| chr9_+_47281961 | 0.10 |
ENSRNOT00000065234
|
Slc9a4
|
solute carrier family 9 member A4 |
| chr20_-_50564987 | 0.10 |
ENSRNOT00000034485
ENSRNOT00000064466 |
Lin28b
|
lin-28 homolog B |
| chr1_+_89220083 | 0.10 |
ENSRNOT00000093144
|
Dmkn
|
dermokine |
| chr13_-_84217332 | 0.10 |
ENSRNOT00000047488
ENSRNOT00000087096 |
Pou2f1
|
POU class 2 homeobox 1 |
| chr1_-_25839198 | 0.10 |
ENSRNOT00000090388
ENSRNOT00000092757 ENSRNOT00000042072 |
Trdn
|
triadin |
| chr2_+_242882306 | 0.10 |
ENSRNOT00000013661
|
Ddit4l
|
DNA-damage-inducible transcript 4-like |
| chr10_-_21265026 | 0.10 |
ENSRNOT00000011922
|
Tenm2
|
teneurin transmembrane protein 2 |
| chr3_-_158328881 | 0.10 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chr9_+_61738471 | 0.10 |
ENSRNOT00000090305
|
AABR07067762.1
|
|
| chr2_+_69415057 | 0.10 |
ENSRNOT00000013152
|
Cdh10
|
cadherin 10 |
| chr8_-_96547568 | 0.10 |
ENSRNOT00000078343
|
RGD1560775
|
similar to RIKEN cDNA 4930579C12 gene |
| chr17_-_43675934 | 0.10 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr13_+_57243877 | 0.09 |
ENSRNOT00000083693
|
Kcnt2
|
potassium sodium-activated channel subfamily T member 2 |
| chr3_-_26056818 | 0.09 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chrX_+_120901495 | 0.09 |
ENSRNOT00000040742
|
Wdr44
|
WD repeat domain 44 |
| chr11_+_82194657 | 0.09 |
ENSRNOT00000002445
|
Etv5
|
ets variant 5 |
| chr16_-_85306366 | 0.09 |
ENSRNOT00000089650
|
Tnfsf13b
|
tumor necrosis factor superfamily member 13b |
| chr13_-_74923402 | 0.09 |
ENSRNOT00000033324
|
Rasal2
|
RAS protein activator like 2 |
| chr1_-_16203838 | 0.09 |
ENSRNOT00000018556
ENSRNOT00000078586 |
Pde7b
|
phosphodiesterase 7B |
| chr9_-_88816898 | 0.09 |
ENSRNOT00000083667
|
Slc19a3
|
solute carrier family 19 member 3 |
| chr5_-_57947183 | 0.08 |
ENSRNOT00000060642
|
Fam219a
|
family with sequence similarity 219, member A |
| chr7_-_140506925 | 0.08 |
ENSRNOT00000083354
|
Kmt2d
|
lysine methyltransferase 2D |
| chr7_-_104800536 | 0.08 |
ENSRNOT00000085650
|
Asap1
|
ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 |
| chr18_-_36322320 | 0.08 |
ENSRNOT00000060260
|
Grxcr2
|
glutaredoxin and cysteine rich domain containing 2 |
| chr5_-_104980641 | 0.08 |
ENSRNOT00000071192
|
Haus6
|
HAUS augmin-like complex, subunit 6 |
| chr17_+_49417067 | 0.08 |
ENSRNOT00000090024
|
Pou6f2
|
POU domain, class 6, transcription factor 2 |
| chr5_-_147867583 | 0.08 |
ENSRNOT00000000139
|
Kpna6
|
karyopherin subunit alpha 6 |
| chr14_+_42007312 | 0.08 |
ENSRNOT00000063985
|
Atp8a1
|
ATPase phospholipid transporting 8A1 |
| chr19_-_22194740 | 0.08 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr1_-_6970040 | 0.08 |
ENSRNOT00000016273
|
Utrn
|
utrophin |
| chr2_+_123597340 | 0.08 |
ENSRNOT00000058552
|
RGD1307100
|
similar to RIKEN cDNA D630029K19 |
| chr5_-_153840178 | 0.07 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chrX_+_24891156 | 0.07 |
ENSRNOT00000071173
|
Wwc3
|
WWC family member 3 |
| chr18_+_24540659 | 0.07 |
ENSRNOT00000061074
|
Ammecr1l
|
AMMECR1 like |
| chr2_-_258997138 | 0.07 |
ENSRNOT00000045020
ENSRNOT00000042256 |
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr12_+_21981755 | 0.07 |
ENSRNOT00000001871
|
LOC100910540
|
7SK snRNA methylphosphate capping enzyme-like |
| chr3_-_71798531 | 0.07 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr6_+_7900972 | 0.07 |
ENSRNOT00000006953
ENSRNOT00000068030 |
Dync2li1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr2_+_113984646 | 0.07 |
ENSRNOT00000016799
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr1_+_101603222 | 0.07 |
ENSRNOT00000033278
|
Izumo1
|
izumo sperm-egg fusion 1 |
| chr3_-_66885085 | 0.07 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr10_-_70802782 | 0.06 |
ENSRNOT00000045867
|
Ccl6
|
chemokine (C-C motif) ligand 6 |
| chr12_+_37878653 | 0.06 |
ENSRNOT00000091084
|
Pitpnm2
|
phosphatidylinositol transfer protein, membrane-associated 2 |
| chr15_-_48445588 | 0.06 |
ENSRNOT00000077197
ENSRNOT00000018583 |
Extl3
|
exostosin-like glycosyltransferase 3 |
| chr6_-_30187337 | 0.06 |
ENSRNOT00000074150
|
Itsn2
|
intersectin 2 |
| chr2_-_177950517 | 0.06 |
ENSRNOT00000012792
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr7_+_41475163 | 0.06 |
ENSRNOT00000037844
|
Dusp6
|
dual specificity phosphatase 6 |
| chr10_-_37311625 | 0.06 |
ENSRNOT00000043343
|
Jade2
|
jade family PHD finger 2 |
| chr16_+_21399553 | 0.06 |
ENSRNOT00000034446
|
LOC108348302
|
zinc finger protein 709-like |
| chr9_-_66845403 | 0.06 |
ENSRNOT00000041546
|
Ica1l
|
islet cell autoantigen 1-like |
| chr7_-_50278842 | 0.06 |
ENSRNOT00000088950
|
Syt1
|
synaptotagmin 1 |
| chr15_-_90175802 | 0.06 |
ENSRNOT00000013342
|
Spry2
|
sprouty RTK signaling antagonist 2 |
| chr19_-_34752695 | 0.06 |
ENSRNOT00000052018
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chrX_-_74968405 | 0.06 |
ENSRNOT00000035653
|
RGD1561931
|
similar to KIAA2022 protein |
| chr10_-_36322356 | 0.06 |
ENSRNOT00000000248
|
Zfp354c
|
zinc finger protein 354C |
| chr1_-_222350173 | 0.06 |
ENSRNOT00000030625
|
Flrt1
|
fibronectin leucine rich transmembrane protein 1 |
| chr7_+_40217991 | 0.06 |
ENSRNOT00000085684
|
Cep290
|
centrosomal protein 290 |
| chr9_-_60672246 | 0.06 |
ENSRNOT00000017761
|
Hecw2
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr15_-_104168564 | 0.06 |
ENSRNOT00000093385
ENSRNOT00000038596 |
Dzip1
|
DAZ interacting zinc finger protein 1 |
| chr7_+_11582984 | 0.06 |
ENSRNOT00000026893
|
Gng7
|
G protein subunit gamma 7 |
| chr2_-_186232292 | 0.06 |
ENSRNOT00000087088
|
Dclk2
|
doublecortin-like kinase 2 |
| chr19_+_23389375 | 0.06 |
ENSRNOT00000018629
|
Sall1
|
spalt-like transcription factor 1 |
| chr5_+_116420690 | 0.06 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr16_-_9658484 | 0.06 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr1_-_49844547 | 0.06 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr8_+_99713783 | 0.05 |
ENSRNOT00000089624
|
Plscr1
|
phospholipid scramblase 1 |
| chr2_+_104020955 | 0.05 |
ENSRNOT00000045586
|
Mtfr1
|
mitochondrial fission regulator 1 |
| chr10_-_103619083 | 0.05 |
ENSRNOT00000029092
|
Cd300e
|
Cd300e molecule |
| chr17_-_1648973 | 0.05 |
ENSRNOT00000036212
|
Slc35d2
|
solute carrier family 35 member D2 |
| chr11_-_61287914 | 0.05 |
ENSRNOT00000086286
|
Spice1
|
spindle and centriole associated protein 1 |
| chr5_-_6186329 | 0.05 |
ENSRNOT00000012610
|
Sulf1
|
sulfatase 1 |
| chr8_+_55504359 | 0.05 |
ENSRNOT00000059086
|
Btg4
|
BTG anti-proliferation factor 4 |
| chr11_-_90406797 | 0.05 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr13_-_108178609 | 0.05 |
ENSRNOT00000004525
|
Cenpf
|
centromere protein F |
| chr8_-_58253688 | 0.05 |
ENSRNOT00000010956
|
Cul5
|
cullin 5 |
| chr3_+_100786862 | 0.05 |
ENSRNOT00000080190
|
Bdnf
|
brain-derived neurotrophic factor |
| chr20_+_28078500 | 0.05 |
ENSRNOT00000030005
|
Ccdc138
|
coiled-coil domain containing 138 |
| chr14_+_39964588 | 0.05 |
ENSRNOT00000003240
|
Gabrg1
|
gamma-aminobutyric acid type A receptor gamma 1 subunit |
| chr7_+_141346398 | 0.05 |
ENSRNOT00000077175
|
Asic1
|
acid sensing ion channel subunit 1 |
| chr1_-_4011161 | 0.05 |
ENSRNOT00000044778
|
Stxbp5
|
syntaxin binding protein 5 |
| chr3_+_112428395 | 0.05 |
ENSRNOT00000079109
ENSRNOT00000048141 |
Stard9
|
StAR-related lipid transfer domain containing 9 |
| chr2_-_19808937 | 0.05 |
ENSRNOT00000044237
|
Atp6ap1l
|
ATPase H+ transporting accessory protein 1 like |
| chr6_-_76608864 | 0.05 |
ENSRNOT00000010824
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr6_+_44230985 | 0.05 |
ENSRNOT00000005858
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr14_-_6793558 | 0.05 |
ENSRNOT00000002927
|
Mepe
|
matrix extracellular phosphoglycoprotein |
| chr7_-_101138860 | 0.05 |
ENSRNOT00000077137
|
AABR07058124.4
|
|
| chr3_+_54253949 | 0.04 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr1_-_87147308 | 0.04 |
ENSRNOT00000027773
ENSRNOT00000089305 ENSRNOT00000090402 |
Actn4
|
actinin alpha 4 |
| chr3_+_100787449 | 0.04 |
ENSRNOT00000078543
|
Bdnf
|
brain-derived neurotrophic factor |
| chr3_-_94418711 | 0.04 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chr8_+_115546893 | 0.04 |
ENSRNOT00000018932
|
Dcaf1
|
DDB1 and CUL4 associated factor 1 |
| chr2_+_147496229 | 0.04 |
ENSRNOT00000022105
|
Tm4sf4
|
transmembrane 4 L six family member 4 |
| chr20_+_3176107 | 0.04 |
ENSRNOT00000001036
|
RT1-S3
|
RT1 class Ib, locus S3 |
| chr14_-_92077088 | 0.04 |
ENSRNOT00000085175
|
Grb10
|
growth factor receptor bound protein 10 |
| chr4_+_33890349 | 0.04 |
ENSRNOT00000078680
|
C1galt1
|
core 1 synthase, glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase, 1 |
| chr8_-_94920441 | 0.04 |
ENSRNOT00000014165
|
Cep162
|
centrosomal protein 162 |
| chr2_-_247446882 | 0.04 |
ENSRNOT00000021963
|
Bmpr1b
|
bone morphogenetic protein receptor type 1B |
| chr3_-_160561741 | 0.04 |
ENSRNOT00000018364
|
Kcns1
|
potassium voltage-gated channel, modifier subfamily S, member 1 |
| chr4_-_156629949 | 0.04 |
ENSRNOT00000042249
|
Vom2r52
|
vomeronasal 2 receptor, 52 |
| chr12_-_18487178 | 0.04 |
ENSRNOT00000030757
|
Zfp157
|
zinc finger protein 157 |
| chr5_-_12199283 | 0.04 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr4_+_157523110 | 0.04 |
ENSRNOT00000081640
|
Zfp384
|
zinc finger protein 384 |
| chr6_-_131926272 | 0.04 |
ENSRNOT00000084057
ENSRNOT00000088421 |
Bcl11b
|
B-cell CLL/lymphoma 11B |
| chr7_-_59547174 | 0.04 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr5_-_161875362 | 0.04 |
ENSRNOT00000078237
|
Prdm2
|
PR/SET domain 2 |
| chr3_-_67787990 | 0.04 |
ENSRNOT00000064851
|
Nckap1
|
NCK-associated protein 1 |
| chr7_-_120380200 | 0.04 |
ENSRNOT00000014878
|
RGD1359634
|
similar to RIKEN cDNA 1700088E04 |
| chr1_-_197568165 | 0.04 |
ENSRNOT00000079044
|
Xpo6
|
exportin 6 |
| chr5_+_124442293 | 0.04 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr1_+_219329574 | 0.04 |
ENSRNOT00000071109
|
Cabp2
|
calcium binding protein 2 |
| chr10_-_55586034 | 0.04 |
ENSRNOT00000080769
|
LOC100912917
|
phosphoribosylformylglycinamidine synthase-like |
| chr7_+_26375866 | 0.04 |
ENSRNOT00000059639
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr14_+_7171613 | 0.04 |
ENSRNOT00000081600
|
Klhl8
|
kelch-like family member 8 |
| chr7_-_29986163 | 0.04 |
ENSRNOT00000033123
|
Gas2l3
|
growth arrest-specific 2 like 3 |
| chr6_-_23542928 | 0.04 |
ENSRNOT00000083858
|
Spdya
|
speedy/RINGO cell cycle regulator family member A |
| chr5_-_16799776 | 0.04 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr4_-_163528220 | 0.04 |
ENSRNOT00000090813
|
Klri1
|
killer cell lectin-like receptor family I member 1 |
| chr7_+_133856101 | 0.04 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr4_-_135069970 | 0.03 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr9_+_77320726 | 0.03 |
ENSRNOT00000068450
|
Spag16
|
sperm associated antigen 16 |
| chr12_-_29743705 | 0.03 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr5_+_171274544 | 0.03 |
ENSRNOT00000077307
|
Cep104
|
centrosomal protein 104 |
| chr10_+_46722109 | 0.03 |
ENSRNOT00000032685
|
Drc3
|
dynein regulatory complex subunit 3 |
| chr4_-_117568348 | 0.03 |
ENSRNOT00000071447
|
Nat8f2
|
N-acetyltransferase 8 (GCN5-related) family member 2 |
| chr15_+_4850122 | 0.03 |
ENSRNOT00000071133
|
Gng2
|
G protein subunit gamma 2 |
| chr7_-_111722964 | 0.03 |
ENSRNOT00000041802
|
Rps19l1
|
ribosomal protein S19-like 1 |
| chr1_+_161922141 | 0.03 |
ENSRNOT00000078300
ENSRNOT00000015784 |
Nars2
|
asparaginyl-tRNA synthetase 2 (mitochondrial)(putative) |
| chr14_-_66978499 | 0.03 |
ENSRNOT00000081601
|
Slit2
|
slit guidance ligand 2 |
| chr4_+_61814974 | 0.03 |
ENSRNOT00000074951
|
Akr1b10
|
aldo-keto reductase family 1 member B10 |
| chrX_-_56765893 | 0.03 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr3_+_72134731 | 0.03 |
ENSRNOT00000083592
|
Ypel4
|
yippee-like 4 |
| chr17_-_79085076 | 0.03 |
ENSRNOT00000057851
|
Fam171a1
|
family with sequence similarity 171, member A1 |
| chr18_+_30562178 | 0.03 |
ENSRNOT00000040998
|
LOC108348771
|
protocadherin beta-16-like |
| chr11_+_31943119 | 0.03 |
ENSRNOT00000047843
|
Itsn1
|
intersectin 1 |
| chr3_+_66673071 | 0.03 |
ENSRNOT00000034769
|
Ppp1r1c
|
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr10_-_73787083 | 0.03 |
ENSRNOT00000035280
|
Med13
|
mediator complex subunit 13 |
| chr1_+_167051209 | 0.03 |
ENSRNOT00000055249
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr7_+_123043503 | 0.03 |
ENSRNOT00000026258
ENSRNOT00000086355 |
Tef
|
TEF, PAR bZIP transcription factor |
| chr7_-_120919261 | 0.03 |
ENSRNOT00000019612
|
Josd1
|
Josephin domain containing 1 |
| chr1_+_154446156 | 0.03 |
ENSRNOT00000093052
|
Picalm
|
phosphatidylinositol binding clathrin assembly protein |
| chr14_+_114152472 | 0.03 |
ENSRNOT00000042965
|
Rtn4
|
reticulon 4 |
| chr1_+_72692105 | 0.03 |
ENSRNOT00000032439
|
Tmem150b
|
transmembrane protein 150B |
| chr1_+_15642153 | 0.03 |
ENSRNOT00000079845
|
Map7
|
microtubule-associated protein 7 |
| chr5_+_126698582 | 0.03 |
ENSRNOT00000012427
|
Cdcp2
|
CUB domain containing protein 2 |
| chr14_+_84876781 | 0.03 |
ENSRNOT00000065188
|
Ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr4_-_132740938 | 0.03 |
ENSRNOT00000007185
|
Rybp
|
RING1 and YY1 binding protein |
| chr2_-_104461863 | 0.03 |
ENSRNOT00000016953
|
Crh
|
corticotropin releasing hormone |
| chr8_+_49077053 | 0.03 |
ENSRNOT00000087922
|
Ift46
|
intraflagellar transport 46 |
| chr13_+_98231326 | 0.03 |
ENSRNOT00000003837
|
Cdc42bpa
|
CDC42 binding protein kinase alpha |
| chr8_-_8524643 | 0.03 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr17_-_42422053 | 0.02 |
ENSRNOT00000048298
|
Fam65b
|
family with sequence similarity 65, member B |
| chr1_-_178367828 | 0.02 |
ENSRNOT00000050823
|
AABR07071968.1
|
|
| chrX_+_11648989 | 0.02 |
ENSRNOT00000041003
|
Bcor
|
BCL6 co-repressor |
| chr10_+_34355755 | 0.02 |
ENSRNOT00000042660
|
AC103024.1
|
|
| chr10_-_15465404 | 0.02 |
ENSRNOT00000077826
ENSRNOT00000027593 |
Decr2
|
2,4-dienoyl-CoA reductase 2 |
| chr14_-_108372068 | 0.02 |
ENSRNOT00000088116
ENSRNOT00000091143 ENSRNOT00000085886 |
RGD1305110
|
similar to KIAA1841 protein |
| chr7_+_15785410 | 0.02 |
ENSRNOT00000082664
ENSRNOT00000073235 |
Zfp955a
|
zinc finger protein 955A |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou4f1_Pou6f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.1 | 0.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.2 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.1 | GO:0090158 | endoplasmic reticulum membrane organization(GO:0090158) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.0 | 0.1 | GO:0061092 | regulation of phospholipid translocation(GO:0061091) positive regulation of phospholipid translocation(GO:0061092) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:1901475 | pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0071930 | negative regulation of transcription involved in G1/S transition of mitotic cell cycle(GO:0071930) |
| 0.0 | 0.1 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.1 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0045200 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:2000384 | regulation of ectoderm development(GO:2000383) negative regulation of ectoderm development(GO:2000384) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.2 | GO:0007171 | activation of transmembrane receptor protein tyrosine kinase activity(GO:0007171) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0006933 | negative regulation of cell adhesion involved in substrate-bound cell migration(GO:0006933) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0000012 | single strand break repair(GO:0000012) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.1 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.0 | 0.0 | GO:1990164 | histone H2A phosphorylation(GO:1990164) |
| 0.0 | 0.0 | GO:0002488 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway(GO:0002488) antigen processing and presentation of endogenous peptide antigen via MHC class Ib via ER pathway, TAP-dependent(GO:0002489) TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.0 | GO:0016267 | O-glycan processing, core 1(GO:0016267) |
| 0.0 | 0.1 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.0 | 0.1 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.0 | 0.0 | GO:0009397 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.1 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.0 | 0.1 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.0 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0061193 | taste bud development(GO:0061193) |
| 0.0 | 0.2 | GO:0048563 | post-embryonic organ morphogenesis(GO:0048563) |
| 0.0 | 0.0 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.0 | GO:0071623 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) |
| 0.0 | 0.1 | GO:0051387 | negative regulation of neurotrophin TRK receptor signaling pathway(GO:0051387) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.1 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.0 | 0.0 | GO:0032398 | MHC class Ib protein complex(GO:0032398) |
| 0.0 | 0.2 | GO:0060091 | kinocilium(GO:0060091) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.1 | GO:0042584 | chromaffin granule membrane(GO:0042584) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.1 | GO:0015563 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.1 | GO:0030292 | protein tyrosine kinase inhibitor activity(GO:0030292) |
| 0.0 | 0.2 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.0 | 0.1 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.0 | GO:0016263 | glycoprotein-N-acetylgalactosamine 3-beta-galactosyltransferase activity(GO:0016263) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0035612 | AP-2 adaptor complex binding(GO:0035612) |
| 0.0 | 0.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |