Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
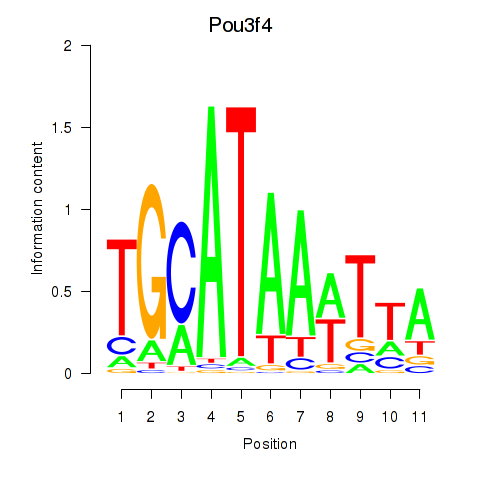
Results for Pou3f4
Z-value: 0.46
Transcription factors associated with Pou3f4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f4
|
ENSRNOG00000002784 | POU class 3 homeobox 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f4 | rn6_v1_chrX_+_82143789_82143789 | 0.85 | 6.7e-02 | Click! |
Activity profile of Pou3f4 motif
Sorted Z-values of Pou3f4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_124929025 | 0.16 |
ENSRNOT00000015447
|
Efcab6
|
EF-hand calcium binding domain 6 |
| chr6_+_106052212 | 0.15 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr2_-_104461863 | 0.14 |
ENSRNOT00000016953
|
Crh
|
corticotropin releasing hormone |
| chr5_-_153840178 | 0.13 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chr6_+_29518416 | 0.12 |
ENSRNOT00000078660
|
AABR07063346.1
|
|
| chr1_+_151439409 | 0.11 |
ENSRNOT00000022060
ENSRNOT00000050639 |
Grm5
|
glutamate metabotropic receptor 5 |
| chr7_-_93502571 | 0.11 |
ENSRNOT00000077033
ENSRNOT00000076080 |
Samd12
|
sterile alpha motif domain containing 12 |
| chr7_+_120923274 | 0.11 |
ENSRNOT00000049247
|
Gtpbp1
|
GTP binding protein 1 |
| chr16_-_81880502 | 0.11 |
ENSRNOT00000079096
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr2_+_22909569 | 0.11 |
ENSRNOT00000073871
|
Homer1
|
homer scaffolding protein 1 |
| chr7_-_120518653 | 0.11 |
ENSRNOT00000016362
|
Baiap2l2
|
BAI1-associated protein 2-like 2 |
| chr14_-_55081551 | 0.11 |
ENSRNOT00000049245
|
Pcdh7
|
protocadherin 7 |
| chr3_+_162357356 | 0.10 |
ENSRNOT00000030119
|
Eya2
|
EYA transcriptional coactivator and phosphatase 2 |
| chr5_-_161875362 | 0.10 |
ENSRNOT00000078237
|
Prdm2
|
PR/SET domain 2 |
| chr3_+_8928534 | 0.10 |
ENSRNOT00000088509
ENSRNOT00000065300 |
Miga2
|
mitoguardin 2 |
| chr1_+_217151166 | 0.10 |
ENSRNOT00000071484
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr10_-_67285617 | 0.10 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chr1_+_167051209 | 0.10 |
ENSRNOT00000055249
|
Numa1
|
nuclear mitotic apparatus protein 1 |
| chr3_-_107760550 | 0.09 |
ENSRNOT00000077091
ENSRNOT00000051638 |
Meis2
|
Meis homeobox 2 |
| chr9_+_119517101 | 0.09 |
ENSRNOT00000020476
|
Lpin2
|
lipin 2 |
| chr11_-_90406797 | 0.09 |
ENSRNOT00000073049
|
Snai2
|
snail family transcriptional repressor 2 |
| chr3_+_72540538 | 0.09 |
ENSRNOT00000012379
|
Aplnr
|
apelin receptor |
| chr1_-_49844547 | 0.09 |
ENSRNOT00000086127
ENSRNOT00000077423 ENSRNOT00000089439 ENSRNOT00000090521 |
AABR07001519.1
|
|
| chr11_+_82194657 | 0.09 |
ENSRNOT00000002445
|
Etv5
|
ets variant 5 |
| chr7_+_121408829 | 0.09 |
ENSRNOT00000023434
|
Mgat3
|
mannosyl (beta-1,4-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase |
| chr6_+_44230985 | 0.09 |
ENSRNOT00000005858
|
Kidins220
|
kinase D-interacting substrate 220 |
| chr1_+_156552328 | 0.09 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr9_+_2190915 | 0.08 |
ENSRNOT00000077417
|
Satb1
|
SATB homeobox 1 |
| chr16_-_9658484 | 0.08 |
ENSRNOT00000065216
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr1_-_88558696 | 0.08 |
ENSRNOT00000038642
|
AABR07002896.1
|
|
| chr7_+_133400485 | 0.08 |
ENSRNOT00000006219
|
Cntn1
|
contactin 1 |
| chr5_+_116420690 | 0.08 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr10_+_54155876 | 0.08 |
ENSRNOT00000072855
|
Gas7
|
growth arrest specific 7 |
| chr20_-_31598118 | 0.07 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr10_+_71536533 | 0.07 |
ENSRNOT00000088138
|
Acaca
|
acetyl-CoA carboxylase alpha |
| chr3_-_94418711 | 0.07 |
ENSRNOT00000089554
|
Hipk3
|
homeodomain interacting protein kinase 3 |
| chrX_+_88298266 | 0.07 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr5_+_27326762 | 0.07 |
ENSRNOT00000066191
|
Runx1t1
|
RUNX1 translocation partner 1 |
| chr18_-_70924708 | 0.07 |
ENSRNOT00000025257
|
Lipg
|
lipase G, endothelial type |
| chr5_+_145257714 | 0.06 |
ENSRNOT00000019214
|
Dlgap3
|
DLG associated protein 3 |
| chr1_+_65781547 | 0.06 |
ENSRNOT00000026204
|
Zfp329
|
zinc finger protein 329 |
| chr17_-_43675934 | 0.06 |
ENSRNOT00000081345
|
Hist1h1t
|
histone cluster 1 H1 family member t |
| chr2_-_147693082 | 0.06 |
ENSRNOT00000022507
|
Wwtr1
|
WW domain containing transcription regulator 1 |
| chr15_-_55424024 | 0.06 |
ENSRNOT00000077733
|
Nudt15
|
nudix hydrolase 15 |
| chr6_+_123034304 | 0.06 |
ENSRNOT00000078938
|
Cpg1
|
candidate plasticity gene 1 |
| chr1_-_6970040 | 0.05 |
ENSRNOT00000016273
|
Utrn
|
utrophin |
| chr18_+_30527705 | 0.05 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr8_-_63291966 | 0.05 |
ENSRNOT00000077762
|
AABR07070278.1
|
|
| chr8_+_82043403 | 0.05 |
ENSRNOT00000091789
|
Myo5a
|
myosin VA |
| chr18_+_30398113 | 0.05 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr1_+_236031988 | 0.05 |
ENSRNOT00000016164
|
Pcsk5
|
proprotein convertase subtilisin/kexin type 5 |
| chr11_+_20474483 | 0.05 |
ENSRNOT00000082417
ENSRNOT00000002895 |
Ncam2
|
neural cell adhesion molecule 2 |
| chr3_-_150960030 | 0.05 |
ENSRNOT00000024714
|
Ncoa6
|
nuclear receptor coactivator 6 |
| chr7_+_28956512 | 0.05 |
ENSRNOT00000006946
|
Gnptab
|
N-acetylglucosamine-1-phosphate transferase, alpha and beta subunits |
| chr5_-_12199283 | 0.05 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr5_-_57947183 | 0.05 |
ENSRNOT00000060642
|
Fam219a
|
family with sequence similarity 219, member A |
| chr12_-_22417980 | 0.05 |
ENSRNOT00000072208
|
Ephb4
|
EPH receptor B4 |
| chrX_+_78042859 | 0.04 |
ENSRNOT00000003286
|
Lpar4
|
lysophosphatidic acid receptor 4 |
| chr13_-_83425641 | 0.04 |
ENSRNOT00000063870
|
Tbx19
|
T-box 19 |
| chr12_-_9607168 | 0.04 |
ENSRNOT00000001260
|
Gsx1
|
GS homeobox 1 |
| chr15_+_12827707 | 0.04 |
ENSRNOT00000012452
|
Fezf2
|
Fez family zinc finger 2 |
| chr11_+_42259761 | 0.04 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chrX_+_11648989 | 0.04 |
ENSRNOT00000041003
|
Bcor
|
BCL6 co-repressor |
| chr16_+_32457521 | 0.04 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr2_+_12516702 | 0.04 |
ENSRNOT00000050072
|
Tmem161b
|
transmembrane protein 161B |
| chr6_-_135251073 | 0.04 |
ENSRNOT00000088986
|
Mok
|
MOK protein kinase |
| chr1_+_127604197 | 0.04 |
ENSRNOT00000018463
|
Lins1
|
lines homolog 1 |
| chr2_+_41467064 | 0.04 |
ENSRNOT00000073231
|
AABR07008066.2
|
|
| chr1_-_166988062 | 0.04 |
ENSRNOT00000039564
|
Lrtomt
|
leucine rich transmembrane and O-methyltransferase domain containing |
| chr2_+_182723854 | 0.04 |
ENSRNOT00000012658
|
Sfrp2
|
secreted frizzled-related protein 2 |
| chr17_+_26478315 | 0.03 |
ENSRNOT00000021669
|
Slc35b3
|
solute carrier family 35 member B3 |
| chr4_-_135069970 | 0.03 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr11_+_31640407 | 0.03 |
ENSRNOT00000029985
ENSRNOT00000091472 |
Ifnar1
|
interferon alpha and beta receptor subunit 1 |
| chr5_-_16799776 | 0.03 |
ENSRNOT00000011721
|
Plag1
|
PLAG1 zinc finger |
| chr4_+_157125998 | 0.03 |
ENSRNOT00000078349
|
C1r
|
complement C1r |
| chr5_+_63870432 | 0.03 |
ENSRNOT00000007641
|
Stx17
|
syntaxin 17 |
| chr12_-_29743705 | 0.03 |
ENSRNOT00000001185
|
Caln1
|
calneuron 1 |
| chr1_-_251387002 | 0.03 |
ENSRNOT00000092063
|
Atad1
|
ATPase family, AAA domain containing 1 |
| chr7_-_140506925 | 0.03 |
ENSRNOT00000083354
|
Kmt2d
|
lysine methyltransferase 2D |
| chr2_+_72006099 | 0.03 |
ENSRNOT00000034044
|
Cdh12
|
cadherin 12 |
| chr9_-_81772851 | 0.02 |
ENSRNOT00000032719
|
Usp37
|
ubiquitin specific peptidase 37 |
| chr5_-_58198782 | 0.02 |
ENSRNOT00000023951
|
Ccl21
|
C-C motif chemokine ligand 21 |
| chr7_-_13108630 | 0.02 |
ENSRNOT00000087038
ENSRNOT00000080491 |
Giot1
|
gonadotropin inducible ovarian transcription factor 1 |
| chr2_-_2779109 | 0.02 |
ENSRNOT00000017098
|
Rfesd
|
Rieske (Fe-S) domain containing |
| chr10_-_88842233 | 0.02 |
ENSRNOT00000026760
|
Stat3
|
signal transducer and activator of transcription 3 |
| chr6_+_102215603 | 0.02 |
ENSRNOT00000014257
|
Plekhh1
|
pleckstrin homology, MyTH4 and FERM domain containing H1 |
| chr12_+_21981755 | 0.02 |
ENSRNOT00000001871
|
LOC100910540
|
7SK snRNA methylphosphate capping enzyme-like |
| chr9_+_19451630 | 0.02 |
ENSRNOT00000065048
|
Enpp4
|
ectonucleotide pyrophosphatase/phosphodiesterase 4 |
| chr1_+_174702373 | 0.02 |
ENSRNOT00000013733
|
Zfp143
|
zinc finger protein 143 |
| chr1_+_61733805 | 0.02 |
ENSRNOT00000086029
|
LOC682206
|
similar to Zinc finger protein 208 |
| chr19_+_25120128 | 0.02 |
ENSRNOT00000081243
|
Samd1
|
sterile alpha motif domain containing 1 |
| chr10_-_87300728 | 0.02 |
ENSRNOT00000036023
ENSRNOT00000081709 |
Krt10
|
keratin 10 |
| chr3_-_26056818 | 0.02 |
ENSRNOT00000044209
|
Lrp1b
|
LDL receptor related protein 1B |
| chr18_-_36322320 | 0.02 |
ENSRNOT00000060260
|
Grxcr2
|
glutaredoxin and cysteine rich domain containing 2 |
| chr20_+_25990304 | 0.02 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr6_-_50941248 | 0.02 |
ENSRNOT00000084533
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr6_-_18351536 | 0.01 |
ENSRNOT00000061588
|
Crnkl1
|
crooked neck pre-mRNA splicing factor 1 |
| chr17_+_1305016 | 0.01 |
ENSRNOT00000025965
|
Ercc6l2
|
ERCC excision repair 6 like 2 |
| chr5_+_48313599 | 0.01 |
ENSRNOT00000081825
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr15_-_71779033 | 0.01 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr10_-_87335823 | 0.01 |
ENSRNOT00000079297
|
Krt12
|
keratin 12 |
| chr10_-_13446135 | 0.01 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr14_+_114152472 | 0.01 |
ENSRNOT00000042965
|
Rtn4
|
reticulon 4 |
| chr19_-_11669578 | 0.01 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chr6_+_7900972 | 0.01 |
ENSRNOT00000006953
ENSRNOT00000068030 |
Dync2li1
|
dynein cytoplasmic 2 light intermediate chain 1 |
| chr5_+_126698582 | 0.01 |
ENSRNOT00000012427
|
Cdcp2
|
CUB domain containing protein 2 |
| chr16_+_9486832 | 0.00 |
ENSRNOT00000082208
|
Arhgap22
|
Rho GTPase activating protein 22 |
| chr5_-_156689415 | 0.00 |
ENSRNOT00000083474
|
Pink1
|
PTEN induced putative kinase 1 |
| chr11_-_62128044 | 0.00 |
ENSRNOT00000093141
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr16_-_48921242 | 0.00 |
ENSRNOT00000041596
|
Cenpu
|
centromere protein U |
| chr13_-_82295123 | 0.00 |
ENSRNOT00000090120
|
LOC498265
|
similar to hypothetical protein FLJ10706 |
| chr1_+_219329574 | 0.00 |
ENSRNOT00000071109
|
Cabp2
|
calcium binding protein 2 |
| chr17_-_14679409 | 0.00 |
ENSRNOT00000020704
|
Aspn
|
asporin |
| chr9_-_45558993 | 0.00 |
ENSRNOT00000087866
ENSRNOT00000017714 |
Chst10
|
carbohydrate sulfotransferase 10 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) negative regulation of glucagon secretion(GO:0070093) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0044268 | multicellular organismal protein metabolic process(GO:0044268) malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.1 | GO:0042078 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.0 | GO:0065001 | specification of axis polarity(GO:0065001) |
| 0.0 | 0.1 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.0 | GO:2000040 | sclerotome development(GO:0061056) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of planar cell polarity pathway involved in axis elongation(GO:2000040) negative regulation of planar cell polarity pathway involved in axis elongation(GO:2000041) |
| 0.0 | 0.1 | GO:0042262 | dGTP catabolic process(GO:0006203) DNA protection(GO:0042262) |
| 0.0 | 0.1 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.0 | GO:2000523 | regulation of aorta morphogenesis(GO:1903847) positive regulation of aorta morphogenesis(GO:1903849) regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.0 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0045203 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
| 0.0 | 0.1 | GO:0061673 | mitotic spindle astral microtubule(GO:0061673) |
| 0.0 | 0.1 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0071439 | clathrin complex(GO:0071439) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0051431 | corticotropin-releasing hormone receptor 2 binding(GO:0051431) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0031802 | type 5 metabotropic glutamate receptor binding(GO:0031802) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.0 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.0 | 0.0 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.0 | GO:0070915 | lysophosphatidic acid binding(GO:0035727) lysophosphatidic acid receptor activity(GO:0070915) |