Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
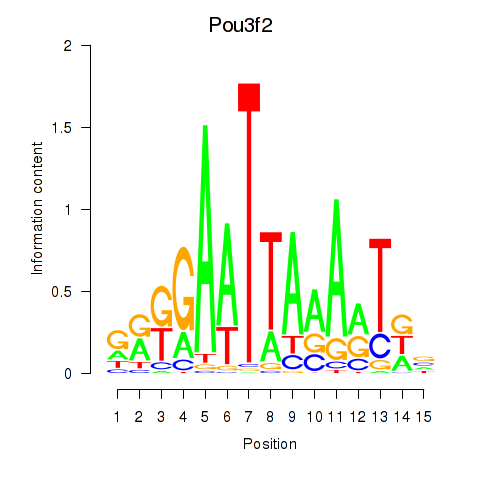
Results for Pou3f2
Z-value: 0.27
Transcription factors associated with Pou3f2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou3f2
|
ENSRNOG00000006908 | POU class 3 homeobox 2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou3f2 | rn6_v1_chr5_-_36683356_36683356 | 0.36 | 5.5e-01 | Click! |
Activity profile of Pou3f2 motif
Sorted Z-values of Pou3f2 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_127604197 | 0.08 |
ENSRNOT00000018463
|
Lins1
|
lines homolog 1 |
| chr2_-_140618405 | 0.08 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr9_+_119542328 | 0.07 |
ENSRNOT00000082005
|
Lpin2
|
lipin 2 |
| chr13_+_29839867 | 0.07 |
ENSRNOT00000090623
|
AABR07020537.1
|
|
| chr5_+_64053946 | 0.07 |
ENSRNOT00000011622
|
Invs
|
inversin |
| chr5_-_172424081 | 0.06 |
ENSRNOT00000019096
|
Plch2
|
phospholipase C, eta 2 |
| chrX_+_54062935 | 0.06 |
ENSRNOT00000004854
|
Tab3
|
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr8_-_55402711 | 0.06 |
ENSRNOT00000091732
|
Sik2
|
salt-inducible kinase 2 |
| chr8_-_124399494 | 0.06 |
ENSRNOT00000037883
|
Tgfbr2
|
transforming growth factor, beta receptor 2 |
| chr16_-_32753278 | 0.06 |
ENSRNOT00000015759
|
Mfap3l
|
microfibrillar-associated protein 3-like |
| chr9_-_69878706 | 0.06 |
ENSRNOT00000035879
|
Ino80d
|
INO80 complex subunit D |
| chr9_-_110225486 | 0.06 |
ENSRNOT00000045096
|
Efna5
|
ephrin A5 |
| chr3_+_54253949 | 0.06 |
ENSRNOT00000010018
|
B3galt1
|
Beta-1,3-galactosyltransferase 1 |
| chr14_-_71973419 | 0.06 |
ENSRNOT00000079895
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr12_+_18931477 | 0.06 |
ENSRNOT00000077087
|
Spry3
|
sprouty RTK signaling antagonist 3 |
| chr10_-_52290657 | 0.05 |
ENSRNOT00000005293
|
Map2k4
|
mitogen activated protein kinase kinase 4 |
| chr4_-_117268178 | 0.05 |
ENSRNOT00000043201
ENSRNOT00000084049 |
Fbxo41
|
F-box protein 41 |
| chr6_+_73553210 | 0.05 |
ENSRNOT00000006562
|
Akap6
|
A-kinase anchoring protein 6 |
| chr14_+_71542057 | 0.05 |
ENSRNOT00000082592
ENSRNOT00000083701 ENSRNOT00000084322 |
Prom1
|
prominin 1 |
| chr15_+_83442144 | 0.05 |
ENSRNOT00000039344
|
Bora
|
bora, aurora kinase A activator |
| chr11_-_11213821 | 0.05 |
ENSRNOT00000093706
|
Robo2
|
roundabout guidance receptor 2 |
| chr12_-_5773036 | 0.05 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr8_-_114010277 | 0.05 |
ENSRNOT00000045087
|
Atp2c1
|
ATPase secretory pathway Ca2+ transporting 1 |
| chr13_+_24823488 | 0.05 |
ENSRNOT00000019907
|
Cdh20
|
cadherin 20 |
| chr10_-_58771908 | 0.05 |
ENSRNOT00000018808
|
RGD1304728
|
similar to 4933427D14Rik protein |
| chr19_-_17399548 | 0.05 |
ENSRNOT00000075991
|
Chd9
|
chromodomain helicase DNA binding protein 9 |
| chr6_+_9790422 | 0.05 |
ENSRNOT00000020959
|
Prkce
|
protein kinase C, epsilon |
| chr4_-_125929002 | 0.04 |
ENSRNOT00000083271
|
Magi1
|
membrane associated guanylate kinase, WW and PDZ domain containing 1 |
| chr15_+_61826711 | 0.04 |
ENSRNOT00000068269
|
Elf1
|
E74-like factor 1 |
| chr20_+_27954433 | 0.04 |
ENSRNOT00000064288
|
Lims1
|
LIM zinc finger domain containing 1 |
| chr13_-_74520634 | 0.04 |
ENSRNOT00000077169
|
Ralgps2
|
Ral GEF with PH domain and SH3 binding motif 2 |
| chr17_+_53231343 | 0.04 |
ENSRNOT00000021703
|
Hecw1
|
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr4_-_168517177 | 0.04 |
ENSRNOT00000009151
|
Dusp16
|
dual specificity phosphatase 16 |
| chr15_+_40665041 | 0.04 |
ENSRNOT00000018300
|
Amer2
|
APC membrane recruitment protein 2 |
| chr1_+_213583606 | 0.04 |
ENSRNOT00000088899
|
AC109844.1
|
|
| chr6_+_126874193 | 0.04 |
ENSRNOT00000011775
|
Unc79
|
unc-79 homolog (C. elegans) |
| chrX_-_76708878 | 0.04 |
ENSRNOT00000045534
|
Atrx
|
ATRX, chromatin remodeler |
| chr9_+_62291405 | 0.04 |
ENSRNOT00000051463
|
Plcl1
|
phospholipase C-like 1 |
| chr5_+_24410863 | 0.03 |
ENSRNOT00000010591
|
Tp53inp1
|
tumor protein p53 inducible nuclear protein 1 |
| chr15_-_28104206 | 0.03 |
ENSRNOT00000032536
|
Ang2
|
angiogenin, ribonuclease A family, member 2 |
| chr15_-_83442008 | 0.03 |
ENSRNOT00000039400
|
Mzt1
|
mitotic spindle organizing protein 1 |
| chrX_-_70428364 | 0.03 |
ENSRNOT00000045907
|
P2ry4
|
pyrimidinergic receptor P2Y4 |
| chr13_-_73055631 | 0.03 |
ENSRNOT00000081892
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr19_-_37796089 | 0.03 |
ENSRNOT00000024891
|
Ranbp10
|
RAN binding protein 10 |
| chr1_-_101046208 | 0.03 |
ENSRNOT00000091711
|
Prr12
|
proline rich 12 |
| chr14_+_106153575 | 0.03 |
ENSRNOT00000010264
|
Vps54
|
VPS54 GARP complex subunit |
| chrX_-_75224268 | 0.03 |
ENSRNOT00000089809
|
Abcb7
|
ATP binding cassette subfamily B member 7 |
| chr1_-_90520344 | 0.03 |
ENSRNOT00000078598
|
Kctd15
|
potassium channel tetramerization domain containing 15 |
| chr10_+_17327275 | 0.03 |
ENSRNOT00000005486
|
Ubtd2
|
ubiquitin domain containing 2 |
| chr1_+_266844480 | 0.03 |
ENSRNOT00000027482
|
Taf5
|
TATA-box binding protein associated factor 5 |
| chr15_-_88036354 | 0.03 |
ENSRNOT00000014747
|
Ednrb
|
endothelin receptor type B |
| chr12_+_10063117 | 0.03 |
ENSRNOT00000044038
|
Usp12
|
ubiquitin specific peptidase 12 |
| chr19_-_33080998 | 0.03 |
ENSRNOT00000015985
|
Slc10a7
|
solute carrier family 10, member 7 |
| chr3_-_111994337 | 0.03 |
ENSRNOT00000030502
|
Pla2g4e
|
phospholipase A2, group IVE |
| chr6_+_78567970 | 0.03 |
ENSRNOT00000032743
|
Ttc6
|
tetratricopeptide repeat domain 6 |
| chr10_+_71332803 | 0.03 |
ENSRNOT00000087428
|
Synrg
|
synergin, gamma |
| chr7_-_132143470 | 0.03 |
ENSRNOT00000038946
ENSRNOT00000044092 ENSRNOT00000066528 ENSRNOT00000045553 ENSRNOT00000046744 ENSRNOT00000055742 |
Kif21a
|
kinesin family member 21A |
| chr9_-_32019205 | 0.03 |
ENSRNOT00000016194
|
Adgrb3
|
adhesion G protein-coupled receptor B3 |
| chr11_-_11214141 | 0.03 |
ENSRNOT00000065748
|
Robo2
|
roundabout guidance receptor 2 |
| chr3_-_111266542 | 0.03 |
ENSRNOT00000031672
|
Ino80
|
INO80 complex subunit |
| chr1_+_222746023 | 0.02 |
ENSRNOT00000028787
|
Atl3
|
atlastin GTPase 3 |
| chr10_-_72472675 | 0.02 |
ENSRNOT00000081649
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr14_-_28536260 | 0.02 |
ENSRNOT00000059942
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr5_+_63870432 | 0.02 |
ENSRNOT00000007641
|
Stx17
|
syntaxin 17 |
| chrX_-_56765893 | 0.02 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr18_+_65388685 | 0.02 |
ENSRNOT00000080844
|
Tcf4
|
transcription factor 4 |
| chr12_+_31335372 | 0.02 |
ENSRNOT00000001242
ENSRNOT00000037296 |
Stx2
|
syntaxin 2 |
| chr17_+_8135251 | 0.02 |
ENSRNOT00000016982
|
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr2_-_177950517 | 0.02 |
ENSRNOT00000012792
|
Rapgef2
|
Rap guanine nucleotide exchange factor 2 |
| chr1_+_64740487 | 0.02 |
ENSRNOT00000081213
|
LOC103691005
|
zinc finger protein 679-like |
| chr3_-_151625644 | 0.02 |
ENSRNOT00000026657
|
Rbm12
|
RNA binding motif protein 12 |
| chr15_-_71779033 | 0.02 |
ENSRNOT00000017765
|
Pcdh20
|
protocadherin 20 |
| chr10_-_87153982 | 0.02 |
ENSRNOT00000014414
|
Krt222
|
keratin 222 |
| chr10_-_86645529 | 0.02 |
ENSRNOT00000011947
|
Med24
|
mediator complex subunit 24 |
| chr11_+_66932614 | 0.02 |
ENSRNOT00000003189
|
Slc15a2
|
solute carrier family 15 member 2 |
| chr5_+_60850852 | 0.02 |
ENSRNOT00000016720
|
Trmt10b
|
tRNA methyltransferase 10B |
| chr13_-_99531959 | 0.02 |
ENSRNOT00000005059
|
Wdr26
|
WD repeat domain 26 |
| chr13_-_47916185 | 0.02 |
ENSRNOT00000087793
|
Dyrk3
|
dual specificity tyrosine phosphorylation regulated kinase 3 |
| chr2_-_232178533 | 0.02 |
ENSRNOT00000082173
ENSRNOT00000055607 |
Ap1ar
|
adaptor-related protein complex 1 associated regulatory protein |
| chr7_-_68523364 | 0.02 |
ENSRNOT00000077670
|
Slc16a7
|
solute carrier family 16 member 7 |
| chr4_-_17594598 | 0.02 |
ENSRNOT00000008936
|
Sema3e
|
semaphorin 3E |
| chr17_-_43422846 | 0.02 |
ENSRNOT00000050526
|
Hist1h2aa
|
histone cluster 1 H2A family member a |
| chr3_+_94530586 | 0.02 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr3_-_152159749 | 0.02 |
ENSRNOT00000067490
|
Cpne1
|
copine 1 |
| chr3_+_97723901 | 0.02 |
ENSRNOT00000080416
|
Mpped2
|
metallophosphoesterase domain containing 2 |
| chr15_+_61826937 | 0.02 |
ENSRNOT00000084005
ENSRNOT00000079417 |
Elf1
|
E74-like factor 1 |
| chr15_+_58016238 | 0.02 |
ENSRNOT00000075786
|
Kctd4
|
potassium channel tetramerization domain containing 4 |
| chr6_+_8218696 | 0.02 |
ENSRNOT00000083470
|
Ppm1b
|
protein phosphatase, Mg2+/Mn2+ dependent, 1B |
| chr16_+_86631064 | 0.01 |
ENSRNOT00000019675
|
Efnb2
|
ephrin B2 |
| chr19_+_38846201 | 0.01 |
ENSRNOT00000030768
ENSRNOT00000063977 |
Tango6
|
transport and golgi organization 6 homolog |
| chr3_+_156642464 | 0.01 |
ENSRNOT00000085354
|
Top1
|
topoisomerase (DNA) I |
| chr3_+_128155069 | 0.01 |
ENSRNOT00000051184
ENSRNOT00000006389 |
Plcb1
|
phospholipase C beta 1 |
| chr2_+_3662763 | 0.01 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr19_-_56037077 | 0.01 |
ENSRNOT00000021968
|
Spata2L
|
spermatogenesis associated 2-like |
| chr3_+_60026747 | 0.01 |
ENSRNOT00000081881
|
Scrn3
|
secernin 3 |
| chrX_-_76925195 | 0.01 |
ENSRNOT00000087977
|
Atrx
|
ATRX, chromatin remodeler |
| chr13_+_80125391 | 0.01 |
ENSRNOT00000044190
|
Mir199a2
|
microRNA 199a-2 |
| chr2_-_142235066 | 0.01 |
ENSRNOT00000018385
|
Cog6
|
component of oligomeric golgi complex 6 |
| chr18_-_81682206 | 0.01 |
ENSRNOT00000058219
|
LOC100359752
|
hypothetical protein LOC100359752 |
| chr16_+_23475351 | 0.01 |
ENSRNOT00000068177
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr15_+_18941431 | 0.01 |
ENSRNOT00000092092
|
AABR07017236.1
|
|
| chr17_-_27451832 | 0.01 |
ENSRNOT00000084417
|
Riok1
|
RIO kinase 1 |
| chr2_-_5577369 | 0.01 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr11_+_45401403 | 0.01 |
ENSRNOT00000002240
|
Tbc1d23
|
TBC1 domain family, member 23 |
| chrX_+_113510621 | 0.01 |
ENSRNOT00000025912
|
Nxt2
|
nuclear transport factor 2-like export factor 2 |
| chr4_-_8214555 | 0.01 |
ENSRNOT00000084050
ENSRNOT00000076642 |
Kmt2e
|
lysine methyltransferase 2E |
| chr7_+_66624512 | 0.01 |
ENSRNOT00000079410
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr6_+_91532467 | 0.01 |
ENSRNOT00000082500
ENSRNOT00000064668 |
Klhdc1
|
kelch domain containing 1 |
| chr7_-_124020574 | 0.01 |
ENSRNOT00000055988
|
Poldip3
|
DNA polymerase delta interacting protein 3 |
| chr18_+_3887419 | 0.01 |
ENSRNOT00000093089
|
Lama3
|
laminin subunit alpha 3 |
| chr3_-_48535909 | 0.01 |
ENSRNOT00000008148
|
Fap
|
fibroblast activation protein, alpha |
| chr16_+_54319377 | 0.00 |
ENSRNOT00000090266
|
Mtus1
|
microtubule associated tumor suppressor 1 |
| chr4_-_145329878 | 0.00 |
ENSRNOT00000011827
|
Tada3
|
transcriptional adaptor 3 |
| chr4_+_22898527 | 0.00 |
ENSRNOT00000072455
ENSRNOT00000076123 |
Dbf4
|
DBF4 zinc finger |
| chr5_+_135442031 | 0.00 |
ENSRNOT00000090852
ENSRNOT00000034464 |
Ccdc17
|
coiled-coil domain containing 17 |
| chr17_-_15484269 | 0.00 |
ENSRNOT00000020648
|
Omd
|
osteomodulin |
| chr10_-_56426012 | 0.00 |
ENSRNOT00000092506
ENSRNOT00000084068 |
LOC497938
|
similar to RIKEN cDNA 4933402P03 |
| chr10_-_70181708 | 0.00 |
ENSRNOT00000076598
|
Rffl
|
ring finger and FYVE-like domain containing E3 ubiquitin protein ligase |
| chr5_+_154868534 | 0.00 |
ENSRNOT00000091442
|
Luzp1
|
leucine zipper protein 1 |
| chr4_+_21462779 | 0.00 |
ENSRNOT00000089747
|
Grm3
|
glutamate metabotropic receptor 3 |
| chr20_+_37876650 | 0.00 |
ENSRNOT00000001054
|
Gja1
|
gap junction protein, alpha 1 |
| chr7_-_27552078 | 0.00 |
ENSRNOT00000059538
|
Stab2
|
stabilin 2 |
| chr5_-_73494030 | 0.00 |
ENSRNOT00000022291
|
Actl7b
|
actin-like 7b |
| chr10_+_1920529 | 0.00 |
ENSRNOT00000072296
|
A630010A05Rik
|
RIKEN cDNA A630010A05 gene |
| chr17_-_12669573 | 0.00 |
ENSRNOT00000016942
ENSRNOT00000041726 |
Syk
|
spleen associated tyrosine kinase |
| chr3_+_67538289 | 0.00 |
ENSRNOT00000009839
|
Dnajc10
|
DnaJ heat shock protein family (Hsp40) member C10 |
| chr14_+_45033309 | 0.00 |
ENSRNOT00000002933
|
Tlr6
|
toll-like receptor 6 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou3f2
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.0 | 0.1 | GO:0060971 | embryonic heart tube left/right pattern formation(GO:0060971) |
| 0.0 | 0.0 | GO:0032472 | Golgi calcium ion transport(GO:0032472) |
| 0.0 | 0.0 | GO:1901581 | post-embryonic appendage morphogenesis(GO:0035120) post-embryonic limb morphogenesis(GO:0035127) post-embryonic forelimb morphogenesis(GO:0035128) telomeric repeat-containing RNA transcription(GO:0097393) telomeric repeat-containing RNA transcription from RNA pol II promoter(GO:0097394) regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901580) negative regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901581) positive regulation of telomeric RNA transcription from RNA pol II promoter(GO:1901582) |
| 0.0 | 0.1 | GO:0050925 | negative regulation of negative chemotaxis(GO:0050925) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.1 | GO:0072139 | glomerular parietal epithelial cell differentiation(GO:0072139) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.0 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.0 | GO:0072703 | myofibroblast differentiation(GO:0036446) response to methyl methanesulfonate(GO:0072702) cellular response to methyl methanesulfonate(GO:0072703) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.0 | GO:0033566 | gamma-tubulin complex localization(GO:0033566) |
| 0.0 | 0.0 | GO:0032485 | regulation of Ral protein signal transduction(GO:0032485) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097543 | ciliary inversin compartment(GO:0097543) |
| 0.0 | 0.1 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.0 | GO:1990421 | subtelomeric heterochromatin(GO:1990421) nuclear subtelomeric heterochromatin(GO:1990707) |
| 0.0 | 0.1 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0071914 | prominosome(GO:0071914) |
| 0.0 | 0.0 | GO:0008274 | gamma-tubulin large complex(GO:0000931) gamma-tubulin ring complex(GO:0008274) gamma-tubulin small complex(GO:0008275) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034714 | type III transforming growth factor beta receptor binding(GO:0034714) |
| 0.0 | 0.1 | GO:0008499 | UDP-galactose:beta-N-acetylglucosamine beta-1,3-galactosyltransferase activity(GO:0008499) |
| 0.0 | 0.0 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.0 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |