Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
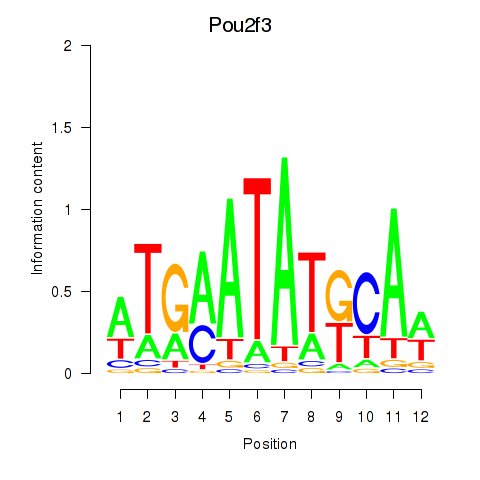
Results for Pou2f3
Z-value: 0.29
Transcription factors associated with Pou2f3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f3
|
ENSRNOG00000009118 | POU class 2 homeobox 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f3 | rn6_v1_chr8_-_47494982_47494982 | -0.10 | 8.7e-01 | Click! |
Activity profile of Pou2f3 motif
Sorted Z-values of Pou2f3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_190370499 | 0.19 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr14_-_6679878 | 0.16 |
ENSRNOT00000075989
ENSRNOT00000067875 |
Spp1
|
secreted phosphoprotein 1 |
| chr2_+_122877286 | 0.15 |
ENSRNOT00000033080
|
Arse
|
arylsulfatase E |
| chr10_-_51576376 | 0.15 |
ENSRNOT00000004829
|
Arhgap44
|
Rho GTPase activating protein 44 |
| chr4_+_148139528 | 0.12 |
ENSRNOT00000092594
ENSRNOT00000015620 ENSRNOT00000092613 |
Washc2c
|
WASH complex subunit 2C |
| chr2_+_127525285 | 0.12 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr1_+_27476375 | 0.11 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr16_+_35116440 | 0.10 |
ENSRNOT00000074353
|
AABR07025358.1
|
|
| chr4_-_11610518 | 0.10 |
ENSRNOT00000066643
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2 |
| chr5_-_153840178 | 0.10 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chr7_+_122433099 | 0.09 |
ENSRNOT00000055447
|
Tmsb10
|
thymosin, beta 10 |
| chr18_-_399242 | 0.09 |
ENSRNOT00000045926
|
F8
|
coagulation factor VIII |
| chr1_+_8310577 | 0.09 |
ENSRNOT00000015131
|
Hivep2
|
human immunodeficiency virus type I enhancer binding protein 2 |
| chr15_+_61673560 | 0.09 |
ENSRNOT00000093661
|
Mtrf1
|
mitochondrial translation release factor 1 |
| chr9_-_65736852 | 0.08 |
ENSRNOT00000087458
|
Trak2
|
trafficking kinesin protein 2 |
| chr12_+_52356832 | 0.08 |
ENSRNOT00000050252
|
Fbrsl1
|
fibrosin-like 1 |
| chr8_-_32328819 | 0.07 |
ENSRNOT00000074457
ENSRNOT00000070818 |
Aplp2
|
amyloid beta precursor like protein 2 |
| chr4_+_168615890 | 0.07 |
ENSRNOT00000009324
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr3_-_148312791 | 0.07 |
ENSRNOT00000091419
|
Bcl2l1
|
Bcl2-like 1 |
| chr4_-_40036563 | 0.07 |
ENSRNOT00000084531
|
Tmem168
|
transmembrane protein 168 |
| chr8_-_63750531 | 0.07 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr16_+_78539489 | 0.07 |
ENSRNOT00000057897
|
Csmd1
|
CUB and Sushi multiple domains 1 |
| chr7_-_70552897 | 0.07 |
ENSRNOT00000080594
|
Kif5a
|
kinesin family member 5A |
| chr16_-_75340360 | 0.07 |
ENSRNOT00000018501
|
Defa5
|
defensin alpha 5 |
| chr4_+_87026530 | 0.07 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr9_-_17209220 | 0.06 |
ENSRNOT00000083764
|
Gtpbp2
|
GTP binding protein 2 |
| chr5_-_161875362 | 0.06 |
ENSRNOT00000078237
|
Prdm2
|
PR/SET domain 2 |
| chr6_+_7793735 | 0.06 |
ENSRNOT00000090724
ENSRNOT00000006832 |
Plekhh2
|
pleckstrin homology, MyTH4 and FERM domain containing H2 |
| chr14_-_28967980 | 0.06 |
ENSRNOT00000048175
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr3_-_112876773 | 0.06 |
ENSRNOT00000015086
|
Ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr8_+_61659445 | 0.06 |
ENSRNOT00000023831
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr3_-_148312420 | 0.06 |
ENSRNOT00000047416
ENSRNOT00000081272 |
Bcl2l1
|
Bcl2-like 1 |
| chr16_-_75481115 | 0.06 |
ENSRNOT00000035128
|
Defa7
|
defensin alpha 7 |
| chr6_-_76535517 | 0.05 |
ENSRNOT00000083857
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr13_+_48455923 | 0.05 |
ENSRNOT00000009280
|
Rab7b
|
Rab7b, member RAS oncogene family |
| chr10_-_88712309 | 0.05 |
ENSRNOT00000081247
|
Stat5b
|
signal transducer and activator of transcription 5B |
| chr14_+_76866135 | 0.05 |
ENSRNOT00000037291
|
Zfp518b
|
zinc finger protein 518B |
| chr18_+_29951094 | 0.05 |
ENSRNOT00000027402
|
LOC102553180
|
protocadherin alpha-1-like |
| chr17_+_36690249 | 0.05 |
ENSRNOT00000089476
ENSRNOT00000082140 |
LOC688583
|
similar to High mobility group protein 4 (HMG-4) (High mobility group protein 2a) (HMG-2a) |
| chr11_+_73078982 | 0.05 |
ENSRNOT00000029549
|
Acap2
|
ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 |
| chr1_-_275882444 | 0.05 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr16_-_48921242 | 0.05 |
ENSRNOT00000041596
|
Cenpu
|
centromere protein U |
| chr19_-_22194740 | 0.05 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr9_-_88816898 | 0.05 |
ENSRNOT00000083667
|
Slc19a3
|
solute carrier family 19 member 3 |
| chr10_-_85049331 | 0.05 |
ENSRNOT00000012538
|
Tbx21
|
T-box 21 |
| chr2_+_122368265 | 0.05 |
ENSRNOT00000078321
|
Atp11b
|
ATPase phospholipid transporting 11B (putative) |
| chr1_+_240355149 | 0.05 |
ENSRNOT00000018521
|
Trpm3
|
transient receptor potential cation channel, subfamily M, member 3 |
| chr7_-_59547174 | 0.04 |
ENSRNOT00000085836
|
Cnot2
|
CCR4-NOT transcription complex, subunit 2 |
| chr8_+_82043403 | 0.04 |
ENSRNOT00000091789
|
Myo5a
|
myosin VA |
| chr3_-_138683318 | 0.04 |
ENSRNOT00000029701
|
Dzank1
|
double zinc ribbon and ankyrin repeat domains 1 |
| chr1_+_80293566 | 0.04 |
ENSRNOT00000024246
|
Ercc2
|
ERCC excision repair 2, TFIIH core complex helicase subunit |
| chr11_+_66316606 | 0.04 |
ENSRNOT00000041715
|
Stxbp5l
|
syntaxin binding protein 5-like |
| chr15_-_39742103 | 0.04 |
ENSRNOT00000074587
ENSRNOT00000036781 ENSRNOT00000071919 |
Setdb2
|
SET domain, bifurcated 2 |
| chr1_+_23409408 | 0.04 |
ENSRNOT00000022362
|
Eya4
|
EYA transcriptional coactivator and phosphatase 4 |
| chr17_-_89163113 | 0.04 |
ENSRNOT00000050445
|
LOC100360843
|
ribosomal protein S19-like |
| chr19_-_34752695 | 0.04 |
ENSRNOT00000052018
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr5_+_151436464 | 0.04 |
ENSRNOT00000012463
|
Map3k6
|
mitogen-activated protein kinase kinase kinase 6 |
| chr3_+_79570625 | 0.04 |
ENSRNOT00000078278
ENSRNOT00000009687 |
Fnbp4
|
formin binding protein 4 |
| chr5_-_150653172 | 0.04 |
ENSRNOT00000072028
|
Med18
|
mediator complex subunit 18 |
| chr5_-_128333805 | 0.04 |
ENSRNOT00000037523
|
Zfyve9
|
zinc finger FYVE-type containing 9 |
| chr1_+_217173199 | 0.04 |
ENSRNOT00000075078
|
Shank2
|
SH3 and multiple ankyrin repeat domains 2 |
| chr4_-_117111653 | 0.04 |
ENSRNOT00000057537
|
Sfxn5
|
sideroflexin 5 |
| chr4_+_155088317 | 0.04 |
ENSRNOT00000088728
|
M6pr
|
mannose-6-phosphate receptor, cation dependent |
| chr1_-_90035522 | 0.04 |
ENSRNOT00000028672
|
Uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr1_+_274688580 | 0.03 |
ENSRNOT00000020580
|
Shoc2
|
SHOC2 leucine-rich repeat scaffold protein |
| chr5_+_119727839 | 0.03 |
ENSRNOT00000014354
|
Cachd1
|
cache domain containing 1 |
| chr3_+_58164931 | 0.03 |
ENSRNOT00000002078
|
Dlx1
|
distal-less homeobox 1 |
| chr1_-_164977633 | 0.03 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr8_+_78872102 | 0.03 |
ENSRNOT00000090047
|
Zfp280d
|
zinc finger protein 280D |
| chr10_+_74959285 | 0.03 |
ENSRNOT00000010296
|
Rnf43
|
ring finger protein 43 |
| chr1_+_219403970 | 0.03 |
ENSRNOT00000029607
|
Ptprcap
|
protein tyrosine phosphatase, receptor type, C-associated protein |
| chr7_-_125497691 | 0.03 |
ENSRNOT00000049445
|
AABR07058578.1
|
|
| chr8_-_63092009 | 0.03 |
ENSRNOT00000034300
|
Loxl1
|
lysyl oxidase-like 1 |
| chr16_+_32457521 | 0.03 |
ENSRNOT00000083579
|
Clcn3
|
chloride voltage-gated channel 3 |
| chr1_+_225015616 | 0.03 |
ENSRNOT00000026416
|
Hnrnpul2
|
heterogeneous nuclear ribonucleoprotein U-like 2 |
| chr5_+_156439633 | 0.03 |
ENSRNOT00000090232
|
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr1_-_67134827 | 0.03 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr18_+_30869628 | 0.03 |
ENSRNOT00000060470
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr1_+_81372650 | 0.03 |
ENSRNOT00000088829
|
Zfp428
|
zinc finger protein 428 |
| chr3_+_172155496 | 0.03 |
ENSRNOT00000066279
|
Stx16
|
syntaxin 16 |
| chr6_-_128434183 | 0.03 |
ENSRNOT00000014405
|
Dicer1
|
dicer 1 ribonuclease III |
| chr10_-_62814520 | 0.03 |
ENSRNOT00000019410
|
Ssh2
|
slingshot protein phosphatase 2 |
| chr17_+_84881414 | 0.03 |
ENSRNOT00000034157
|
Mllt10
|
myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| chr14_+_7171613 | 0.03 |
ENSRNOT00000081600
|
Klhl8
|
kelch-like family member 8 |
| chr3_-_150960030 | 0.03 |
ENSRNOT00000024714
|
Ncoa6
|
nuclear receptor coactivator 6 |
| chr9_+_10941613 | 0.03 |
ENSRNOT00000070794
|
Sema6b
|
semaphorin 6B |
| chr19_+_387374 | 0.03 |
ENSRNOT00000075433
|
LOC102547811
|
uncharacterized LOC102547811 |
| chr7_+_26522314 | 0.03 |
ENSRNOT00000011572
|
Slc41a2
|
solute carrier family 41 member 2 |
| chr14_-_34177455 | 0.03 |
ENSRNOT00000059814
ENSRNOT00000091230 |
Exoc1
|
exocyst complex component 1 |
| chr7_+_120923274 | 0.03 |
ENSRNOT00000049247
|
Gtpbp1
|
GTP binding protein 1 |
| chr12_-_41993957 | 0.03 |
ENSRNOT00000001891
|
Rbm19
|
RNA binding motif protein 19 |
| chr6_+_101532518 | 0.03 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr14_+_34389991 | 0.03 |
ENSRNOT00000002953
|
Pdcl2
|
phosducin-like 2 |
| chr8_+_71547468 | 0.03 |
ENSRNOT00000042369
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr9_-_75528644 | 0.03 |
ENSRNOT00000019283
|
Erbb4
|
erb-b2 receptor tyrosine kinase 4 |
| chr7_-_13751271 | 0.03 |
ENSRNOT00000009931
|
Slc1a6
|
solute carrier family 1 member 6 |
| chrX_+_88298266 | 0.03 |
ENSRNOT00000041508
|
AABR07039936.1
|
|
| chr17_+_35435079 | 0.03 |
ENSRNOT00000074800
|
Exoc2
|
exocyst complex component 2 |
| chr5_+_116420690 | 0.03 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr10_-_110274768 | 0.03 |
ENSRNOT00000054931
|
Sectm1a
|
secreted and transmembrane 1A |
| chrX_-_1952111 | 0.03 |
ENSRNOT00000061140
|
Jade3
|
jade family PHD finger 3 |
| chrX_-_75224268 | 0.03 |
ENSRNOT00000089809
|
Abcb7
|
ATP binding cassette subfamily B member 7 |
| chr9_-_80167033 | 0.03 |
ENSRNOT00000023530
|
Igfbp5
|
insulin-like growth factor binding protein 5 |
| chr1_-_7443863 | 0.03 |
ENSRNOT00000088558
|
Phactr2
|
phosphatase and actin regulator 2 |
| chr1_+_171797516 | 0.03 |
ENSRNOT00000088110
|
Ppfibp2
|
PPFIA binding protein 2 |
| chr15_-_36798814 | 0.03 |
ENSRNOT00000065764
|
Cenpj
|
centromere protein J |
| chr3_+_148150698 | 0.02 |
ENSRNOT00000087075
ENSRNOT00000010251 |
Hm13
|
histocompatibility minor 13 |
| chr1_+_57692836 | 0.02 |
ENSRNOT00000083968
ENSRNOT00000019358 |
Chd1
|
chromodomain helicase DNA binding protein 1 |
| chr17_-_43821536 | 0.02 |
ENSRNOT00000072286
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr11_-_80981415 | 0.02 |
ENSRNOT00000002499
ENSRNOT00000002496 |
St6gal1
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| chr10_+_78111050 | 0.02 |
ENSRNOT00000079364
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr7_+_73273985 | 0.02 |
ENSRNOT00000077730
|
Pop1
|
POP1 homolog, ribonuclease P/MRP subunit |
| chr4_+_174692331 | 0.02 |
ENSRNOT00000011770
|
Plekha5
|
pleckstrin homology domain containing A5 |
| chr8_+_103938520 | 0.02 |
ENSRNOT00000067835
|
Gk5
|
glycerol kinase 5 (putative) |
| chr5_+_122390522 | 0.02 |
ENSRNOT00000064567
|
Sgip1
|
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr5_-_139921013 | 0.02 |
ENSRNOT00000079957
|
Smap2
|
small ArfGAP2 |
| chrX_-_45000363 | 0.02 |
ENSRNOT00000035538
|
Cldn34e
|
claudin 34E |
| chr19_-_37916813 | 0.02 |
ENSRNOT00000026585
|
Lcat
|
lecithin cholesterol acyltransferase |
| chr9_+_46657575 | 0.02 |
ENSRNOT00000083322
|
Map4k4
|
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr4_+_147756553 | 0.02 |
ENSRNOT00000086549
ENSRNOT00000014733 |
Ift122
|
intraflagellar transport 122 |
| chr5_+_155934490 | 0.02 |
ENSRNOT00000077933
|
LOC100911993
|
ubiquitin carboxyl-terminal hydrolase 48-like |
| chr8_-_8524643 | 0.02 |
ENSRNOT00000009418
|
Cntn5
|
contactin 5 |
| chr10_+_76280933 | 0.02 |
ENSRNOT00000051823
|
RGD1304870
|
similar to Tceb2 protein |
| chr4_+_40161285 | 0.02 |
ENSRNOT00000050722
|
LOC500035
|
hypothetical protein LOC500035 |
| chr16_+_6806170 | 0.02 |
ENSRNOT00000084713
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr4_-_72143748 | 0.02 |
ENSRNOT00000024428
|
Tcaf1
|
TRPM8 channel-associated factor 1 |
| chr3_-_11102515 | 0.02 |
ENSRNOT00000035580
|
Fam78a
|
family with sequence similarity 78, member A |
| chr4_+_155654911 | 0.02 |
ENSRNOT00000087883
|
Foxj2
|
forkhead box J2 |
| chr2_-_140387505 | 0.02 |
ENSRNOT00000047635
|
Elf2
|
E74-like factor 2 |
| chr16_-_48692476 | 0.02 |
ENSRNOT00000013118
|
Irf2
|
interferon regulatory factor 2 |
| chr18_+_30474947 | 0.02 |
ENSRNOT00000027188
|
Pcdhb9
|
protocadherin beta 9 |
| chr16_+_71242470 | 0.02 |
ENSRNOT00000030489
|
Letm2
|
leucine zipper and EF-hand containing transmembrane protein 2 |
| chr18_+_30424814 | 0.02 |
ENSRNOT00000073425
|
Pcdhb7
|
protocadherin beta 7 |
| chr8_+_32452885 | 0.02 |
ENSRNOT00000010423
|
Prdm10
|
PR/SET domain 10 |
| chr18_+_30509393 | 0.02 |
ENSRNOT00000043846
|
Pcdhb12
|
protocadherin beta 12 |
| chr10_+_30169257 | 0.02 |
ENSRNOT00000006436
|
Rnf145
|
ring finger protein 145 |
| chr17_+_43679411 | 0.02 |
ENSRNOT00000085865
|
AC130391.3
|
|
| chr9_+_52023295 | 0.02 |
ENSRNOT00000004956
|
Col3a1
|
collagen type III alpha 1 chain |
| chr11_+_42259761 | 0.02 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr4_+_61924013 | 0.02 |
ENSRNOT00000090717
|
Bpgm
|
bisphosphoglycerate mutase |
| chr1_+_72399157 | 0.02 |
ENSRNOT00000022349
|
Zfp579
|
zinc finger protein 579 |
| chr3_-_123997277 | 0.02 |
ENSRNOT00000058238
|
RGD1564425
|
similar to Proteasome subunit beta type 3 (Proteasome theta chain) |
| chr5_-_164714145 | 0.02 |
ENSRNOT00000055680
|
LOC100911485
|
mitofusin-2-like |
| chr11_-_88374679 | 0.02 |
ENSRNOT00000002524
|
Top3b
|
topoisomerase (DNA) III beta |
| chr7_-_120380200 | 0.02 |
ENSRNOT00000014878
|
RGD1359634
|
similar to RIKEN cDNA 1700088E04 |
| chr7_+_133856101 | 0.02 |
ENSRNOT00000038686
|
Pdzrn4
|
PDZ domain containing RING finger 4 |
| chr8_-_93390305 | 0.02 |
ENSRNOT00000056930
|
Ibtk
|
inhibitor of Bruton tyrosine kinase |
| chr6_+_29106141 | 0.02 |
ENSRNOT00000084560
|
Atad2b
|
ATPase family, AAA domain containing 2B |
| chr7_+_11295892 | 0.02 |
ENSRNOT00000089004
|
Tjp3
|
tight junction protein 3 |
| chr5_+_74941802 | 0.02 |
ENSRNOT00000015576
|
Akap2
|
A-kinase anchoring protein 2 |
| chr2_+_2694480 | 0.02 |
ENSRNOT00000083565
|
AABR07072671.1
|
|
| chr2_+_193778042 | 0.02 |
ENSRNOT00000090212
|
Tchh
|
trichohyalin |
| chrX_-_63182385 | 0.02 |
ENSRNOT00000076613
|
Zfx
|
zinc finger protein X-linked |
| chr8_-_116993193 | 0.02 |
ENSRNOT00000026327
|
Dag1
|
dystroglycan 1 |
| chr16_+_3293599 | 0.01 |
ENSRNOT00000081999
ENSRNOT00000047118 ENSRNOT00000020427 |
Erc2
|
ELKS/RAB6-interacting/CAST family member 2 |
| chr14_+_33624390 | 0.01 |
ENSRNOT00000002907
|
Aasdh
|
aminoadipate-semialdehyde dehydrogenase |
| chr15_+_57340579 | 0.01 |
ENSRNOT00000015467
|
Zc3h13
|
zinc finger CCCH type containing 13 |
| chr4_-_78074906 | 0.01 |
ENSRNOT00000080654
|
Zfp467
|
zinc finger protein 467 |
| chr1_+_188420461 | 0.01 |
ENSRNOT00000037870
|
Ccp110
|
centriolar coiled-coil protein 110 |
| chr3_-_143192408 | 0.01 |
ENSRNOT00000006885
|
LOC689081
|
similar to cystatin E2 |
| chr17_+_30556884 | 0.01 |
ENSRNOT00000080929
ENSRNOT00000022022 |
Eci2
|
enoyl-CoA delta isomerase 2 |
| chr12_+_4310334 | 0.01 |
ENSRNOT00000041244
|
Vom2r60
|
vomeronasal 2 receptor, 60 |
| chr15_+_56666012 | 0.01 |
ENSRNOT00000013408
|
Htr2a
|
5-hydroxytryptamine receptor 2A |
| chr11_-_61287914 | 0.01 |
ENSRNOT00000086286
|
Spice1
|
spindle and centriole associated protein 1 |
| chr19_-_44101365 | 0.01 |
ENSRNOT00000082182
|
Tmem170a
|
transmembrane protein 170A |
| chr2_+_248178389 | 0.01 |
ENSRNOT00000037339
|
Gbp5
|
guanylate binding protein 5 |
| chr9_-_14599594 | 0.01 |
ENSRNOT00000018138
|
Treml1
|
triggering receptor expressed on myeloid cells-like 1 |
| chr20_+_11114164 | 0.01 |
ENSRNOT00000001602
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr10_+_34489940 | 0.01 |
ENSRNOT00000085975
|
Zfp62
|
zinc finger protein 62 |
| chr6_-_27487284 | 0.01 |
ENSRNOT00000084344
|
Selenoi
|
selenoprotein I |
| chr10_-_13446135 | 0.01 |
ENSRNOT00000084991
|
Kctd5
|
potassium channel tetramerization domain containing 5 |
| chr7_+_44009069 | 0.01 |
ENSRNOT00000005523
|
Mgat4c
|
MGAT4 family, member C |
| chrX_-_63807810 | 0.01 |
ENSRNOT00000084632
|
Maged1
|
MAGE family member D1 |
| chr8_+_61671513 | 0.01 |
ENSRNOT00000091128
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr17_-_90071408 | 0.01 |
ENSRNOT00000071978
|
Pdss1
|
decaprenyl diphosphate synthase subunit 1 |
| chr1_-_22269831 | 0.01 |
ENSRNOT00000081804
|
Stx7
|
syntaxin 7 |
| chr2_+_266317745 | 0.01 |
ENSRNOT00000080305
|
Wls
|
wntless Wnt ligand secretion mediator |
| chr7_+_72599479 | 0.01 |
ENSRNOT00000018963
|
AABR07057460.1
|
|
| chr2_+_251002213 | 0.01 |
ENSRNOT00000080600
|
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr7_-_138483612 | 0.01 |
ENSRNOT00000085620
|
Slc38a4
|
solute carrier family 38, member 4 |
| chr5_+_47546014 | 0.01 |
ENSRNOT00000065882
|
Bach2
|
BTB domain and CNC homolog 2 |
| chr4_+_88328061 | 0.01 |
ENSRNOT00000084775
|
Vom1r87
|
vomeronasal 1 receptor 87 |
| chr18_+_65388685 | 0.01 |
ENSRNOT00000080844
|
Tcf4
|
transcription factor 4 |
| chr3_-_66279155 | 0.01 |
ENSRNOT00000079887
|
Cerkl
|
ceramide kinase-like |
| chr8_-_71533069 | 0.01 |
ENSRNOT00000021863
|
Trip4
|
thyroid hormone receptor interactor 4 |
| chr2_-_118882562 | 0.01 |
ENSRNOT00000058860
|
Kcnmb3
|
potassium calcium-activated channel subfamily M regulatory beta subunit 3 |
| chr5_+_128083118 | 0.01 |
ENSRNOT00000079161
|
Zcchc11
|
zinc finger CCHC-type containing 11 |
| chr14_-_20816588 | 0.01 |
ENSRNOT00000033806
|
Slc4a4
|
solute carrier family 4 member 4 |
| chr5_-_39215102 | 0.01 |
ENSRNOT00000050653
|
Klhl32
|
kelch-like family member 32 |
| chr2_-_120570355 | 0.01 |
ENSRNOT00000088442
|
Fxr1
|
FMR1 autosomal homolog 1 |
| chr8_+_115193146 | 0.01 |
ENSRNOT00000056391
|
Iqcf1
|
IQ motif containing F1 |
| chr10_+_97771264 | 0.01 |
ENSRNOT00000005257
|
Arsg
|
arylsulfatase G |
| chr5_-_70463546 | 0.01 |
ENSRNOT00000043184
|
AC118841.1
|
|
| chr1_+_259739955 | 0.01 |
ENSRNOT00000054719
|
Entpd1
|
ectonucleoside triphosphate diphosphohydrolase 1 |
| chr2_+_188748359 | 0.01 |
ENSRNOT00000028038
|
Shc1
|
SHC adaptor protein 1 |
| chr4_+_88423412 | 0.01 |
ENSRNOT00000040163
|
Vom1r88
|
vomeronasal 1 receptor 88 |
| chr11_+_31539016 | 0.01 |
ENSRNOT00000072856
|
Ifnar2
|
interferon alpha and beta receptor subunit 2 |
| chr1_+_61733805 | 0.01 |
ENSRNOT00000086029
|
LOC682206
|
similar to Zinc finger protein 208 |
| chr1_+_17602281 | 0.01 |
ENSRNOT00000075461
|
LOC103690149
|
jouberin-like |
| chr20_-_6195463 | 0.01 |
ENSRNOT00000092474
|
Pxt1
|
peroxisomal, testis specific 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.1 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.0 | 0.1 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0098939 | dendritic transport of mitochondrion(GO:0098939) anterograde dendritic transport of mitochondrion(GO:0098972) |
| 0.0 | 0.1 | GO:0098971 | anterograde dendritic transport of neurotransmitter receptor complex(GO:0098971) |
| 0.0 | 0.1 | GO:0034499 | late endosome to Golgi transport(GO:0034499) |
| 0.0 | 0.0 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.0 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.1 | GO:0046544 | development of secondary male sexual characteristics(GO:0046544) |
| 0.0 | 0.0 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.0 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) |
| 0.0 | 0.0 | GO:0051643 | endoplasmic reticulum localization(GO:0051643) |
| 0.0 | 0.0 | GO:0010070 | zygote asymmetric cell division(GO:0010070) targeting of mRNA for destruction involved in RNA interference(GO:0030423) regulation of enamel mineralization(GO:0070173) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.0 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
| 0.0 | 0.0 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.2 | GO:0004065 | arylsulfatase activity(GO:0004065) |
| 0.0 | 0.1 | GO:0016149 | translation release factor activity, codon specific(GO:0016149) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.0 | GO:0015563 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.0 | GO:0001226 | RNA polymerase II transcription corepressor binding(GO:0001226) |
| 0.0 | 0.0 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME THE ACTIVATION OF ARYLSULFATASES | Genes involved in The activation of arylsulfatases |
| 0.0 | 0.1 | REACTOME DEFENSINS | Genes involved in Defensins |