Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Pou2f1
Z-value: 0.81
Transcription factors associated with Pou2f1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pou2f1
|
ENSRNOG00000003581 | POU class 2 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pou2f1 | rn6_v1_chr13_-_84217332_84217366 | 0.33 | 5.9e-01 | Click! |
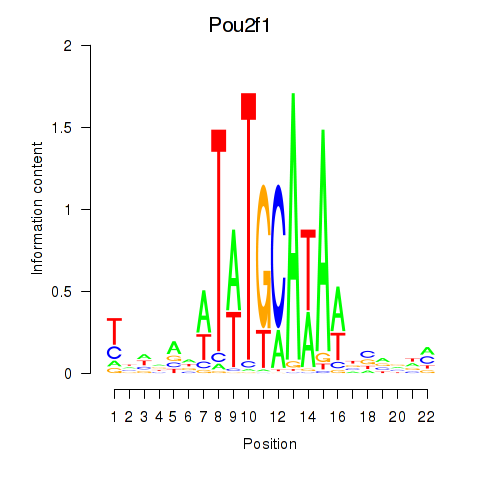
Activity profile of Pou2f1 motif
Sorted Z-values of Pou2f1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_-_126710854 | 0.37 |
ENSRNOT00000081127
ENSRNOT00000064914 |
Btbd7
|
BTB domain containing 7 |
| chr6_-_76535517 | 0.28 |
ENSRNOT00000083857
|
Ralgapa1
|
Ral GTPase activating protein catalytic alpha subunit 1 |
| chr18_+_30890869 | 0.27 |
ENSRNOT00000060466
|
Pcdhgb6
|
protocadherin gamma subfamily B, 6 |
| chr10_+_56381813 | 0.27 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr12_-_38782010 | 0.26 |
ENSRNOT00000001813
|
Wdr66
|
WD repeat domain 66 |
| chr1_-_67134827 | 0.25 |
ENSRNOT00000045214
|
Vom1r45
|
vomeronasal 1 receptor 45 |
| chr5_-_164648328 | 0.25 |
ENSRNOT00000090811
|
Zfp933
|
zinc finger protein 933 |
| chr4_+_94696965 | 0.25 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr7_-_9976367 | 0.24 |
ENSRNOT00000044241
|
Zfp347
|
zinc finger protein 347 |
| chr6_+_64224861 | 0.24 |
ENSRNOT00000093159
ENSRNOT00000093664 |
Pnpla8
|
patatin-like phospholipase domain containing 8 |
| chr2_+_196608496 | 0.23 |
ENSRNOT00000091681
|
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr7_-_13108630 | 0.22 |
ENSRNOT00000087038
ENSRNOT00000080491 |
Giot1
|
gonadotropin inducible ovarian transcription factor 1 |
| chr4_-_22425381 | 0.19 |
ENSRNOT00000011166
ENSRNOT00000037712 |
Abcb1a
|
ATP binding cassette subfamily B member 1A |
| chrX_+_120901495 | 0.19 |
ENSRNOT00000040742
|
Wdr44
|
WD repeat domain 44 |
| chr19_-_34752695 | 0.19 |
ENSRNOT00000052018
|
Nr3c2
|
nuclear receptor subfamily 3, group C, member 2 |
| chr1_-_82344345 | 0.19 |
ENSRNOT00000051892
ENSRNOT00000090629 ENSRNOT00000029487 ENSRNOT00000027933 |
Ceacam1
|
carcinoembryonic antigen related cell adhesion molecule 1 |
| chr4_+_124005280 | 0.18 |
ENSRNOT00000014353
|
Nr2c2
|
nuclear receptor subfamily 2, group C, member 2 |
| chr13_-_25652473 | 0.18 |
ENSRNOT00000020752
|
Pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr16_+_50152008 | 0.18 |
ENSRNOT00000019237
|
Klkb1
|
kallikrein B1 |
| chr15_+_57241968 | 0.17 |
ENSRNOT00000082191
|
Lcp1
|
lymphocyte cytosolic protein 1 |
| chr7_-_58365313 | 0.17 |
ENSRNOT00000005443
|
Thap2
|
THAP domain containing 2 |
| chr8_+_130366775 | 0.17 |
ENSRNOT00000071152
|
Nktr
|
natural killer cell triggering receptor |
| chr6_+_102215603 | 0.17 |
ENSRNOT00000014257
|
Plekhh1
|
pleckstrin homology, MyTH4 and FERM domain containing H1 |
| chr5_-_150167077 | 0.16 |
ENSRNOT00000088493
ENSRNOT00000013761 ENSRNOT00000067349 ENSRNOT00000051097 ENSRNOT00000067189 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr10_-_67285617 | 0.16 |
ENSRNOT00000019044
|
Utp6
|
UTP6 small subunit processome component |
| chr6_-_99783047 | 0.16 |
ENSRNOT00000009028
|
Sptb
|
spectrin, beta, erythrocytic |
| chrX_+_158835811 | 0.15 |
ENSRNOT00000071888
ENSRNOT00000080110 |
Ints6l
|
integrator complex subunit 6 like |
| chr5_-_75319765 | 0.15 |
ENSRNOT00000085698
|
Svep1
|
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr3_-_112876773 | 0.15 |
ENSRNOT00000015086
|
Ubr1
|
ubiquitin protein ligase E3 component n-recognin 1 |
| chr5_-_169658875 | 0.15 |
ENSRNOT00000015840
|
Kcnab2
|
potassium voltage-gated channel subfamily A regulatory beta subunit 2 |
| chr8_+_58347736 | 0.15 |
ENSRNOT00000080227
ENSRNOT00000066222 |
Slc35f2
|
solute carrier family 35, member F2 |
| chr1_+_5448958 | 0.15 |
ENSRNOT00000061930
|
Epm2a
|
epilepsy, progressive myoclonus type 2A |
| chr1_-_164977633 | 0.14 |
ENSRNOT00000029629
|
Rnf169
|
ring finger protein 169 |
| chr4_+_157125998 | 0.14 |
ENSRNOT00000078349
|
C1r
|
complement C1r |
| chr3_-_66279155 | 0.14 |
ENSRNOT00000079887
|
Cerkl
|
ceramide kinase-like |
| chr14_-_16996962 | 0.14 |
ENSRNOT00000091936
|
AC103535.1
|
|
| chr13_-_94289333 | 0.13 |
ENSRNOT00000078195
|
Pld5
|
phospholipase D family, member 5 |
| chr1_-_235405831 | 0.13 |
ENSRNOT00000071578
|
AABR07006458.1
|
|
| chr10_+_40543288 | 0.13 |
ENSRNOT00000016755
|
Slc36a1
|
solute carrier family 36 member 1 |
| chr6_-_54059119 | 0.13 |
ENSRNOT00000005521
|
Hdac9
|
histone deacetylase 9 |
| chr8_+_32452885 | 0.13 |
ENSRNOT00000010423
|
Prdm10
|
PR/SET domain 10 |
| chr5_+_152721940 | 0.12 |
ENSRNOT00000039322
|
Aunip
|
aurora kinase A and ninein interacting protein |
| chr2_+_86891092 | 0.12 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr2_-_165600748 | 0.12 |
ENSRNOT00000013216
|
Ift80
|
intraflagellar transport 80 |
| chr8_+_71547468 | 0.12 |
ENSRNOT00000042369
|
Csnk1g1
|
casein kinase 1, gamma 1 |
| chr2_-_247446882 | 0.12 |
ENSRNOT00000021963
|
Bmpr1b
|
bone morphogenetic protein receptor type 1B |
| chr5_-_117440254 | 0.12 |
ENSRNOT00000066139
|
Kank4
|
KN motif and ankyrin repeat domains 4 |
| chr13_+_43850751 | 0.12 |
ENSRNOT00000004995
|
Mgat5
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr1_-_215846911 | 0.12 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr8_-_102149912 | 0.12 |
ENSRNOT00000011263
|
RGD1309079
|
similar to Ab2-095 |
| chr2_-_86475096 | 0.12 |
ENSRNOT00000075247
ENSRNOT00000075062 |
LOC100909688
|
zinc finger protein 43-like |
| chr7_-_24885775 | 0.12 |
ENSRNOT00000010024
|
Tcp11l2
|
t-complex 11 like 2 |
| chr17_+_78764506 | 0.12 |
ENSRNOT00000077953
|
Suv39h2
|
suppressor of variegation 3-9 homolog 2 |
| chr13_-_35668968 | 0.12 |
ENSRNOT00000042862
ENSRNOT00000003428 |
Epb41l5
|
erythrocyte membrane protein band 4.1 like 5 |
| chr4_-_50693869 | 0.11 |
ENSRNOT00000064228
|
Rnf148
|
ring finger protein 148 |
| chr3_-_66885085 | 0.11 |
ENSRNOT00000084299
ENSRNOT00000090547 |
Pde1a
|
phosphodiesterase 1A |
| chr19_-_914880 | 0.11 |
ENSRNOT00000017127
|
Cklf
|
chemokine-like factor |
| chr1_+_88206813 | 0.11 |
ENSRNOT00000072556
|
LOC103690163
|
zinc finger protein 420 |
| chr8_-_13906355 | 0.11 |
ENSRNOT00000029634
|
Cep295
|
centrosomal protein 295 |
| chr13_+_87986240 | 0.11 |
ENSRNOT00000003705
|
Rgs5
|
regulator of G-protein signaling 5 |
| chr3_-_21669957 | 0.10 |
ENSRNOT00000012354
ENSRNOT00000078718 |
Rc3h2
|
ring finger and CCCH-type domains 2 |
| chr18_+_30424814 | 0.10 |
ENSRNOT00000073425
|
Pcdhb7
|
protocadherin beta 7 |
| chr2_-_116413873 | 0.10 |
ENSRNOT00000010344
|
Mynn
|
myoneurin |
| chr8_+_117737387 | 0.10 |
ENSRNOT00000090164
ENSRNOT00000091573 |
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr11_+_9642365 | 0.10 |
ENSRNOT00000087080
ENSRNOT00000042384 |
Robo1
|
roundabout guidance receptor 1 |
| chr3_-_61488696 | 0.10 |
ENSRNOT00000083045
|
Lnpk
|
lunapark, ER junction formation factor |
| chr7_-_66909470 | 0.10 |
ENSRNOT00000066381
|
Ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chrY_+_506149 | 0.10 |
ENSRNOT00000086056
|
Kdm5d
|
lysine demethylase 5D |
| chr5_+_169213115 | 0.10 |
ENSRNOT00000013535
|
Nol9
|
nucleolar protein 9 |
| chr18_+_29966245 | 0.10 |
ENSRNOT00000074028
|
Pcdha4
|
protocadherin alpha 4 |
| chr2_+_237755673 | 0.10 |
ENSRNOT00000083937
|
Tbck
|
TBC1 domain containing kinase |
| chr18_+_30581530 | 0.10 |
ENSRNOT00000048166
|
Pcdhb20
|
protocadherin beta 20 |
| chr8_-_21481735 | 0.10 |
ENSRNOT00000038069
|
Zfp426
|
zinc finger protein 426 |
| chr10_+_34489940 | 0.09 |
ENSRNOT00000085975
|
Zfp62
|
zinc finger protein 62 |
| chr1_+_87790104 | 0.09 |
ENSRNOT00000074580
|
LOC103690114
|
zinc finger protein 383-like |
| chr11_+_42259761 | 0.09 |
ENSRNOT00000047310
|
Epha6
|
Eph receptor A6 |
| chr13_+_50873605 | 0.09 |
ENSRNOT00000004382
|
Fmod
|
fibromodulin |
| chr11_+_15081774 | 0.09 |
ENSRNOT00000009911
|
LOC108348050
|
ubiquitin carboxyl-terminal hydrolase 25-like |
| chr15_-_39742103 | 0.09 |
ENSRNOT00000074587
ENSRNOT00000036781 ENSRNOT00000071919 |
Setdb2
|
SET domain, bifurcated 2 |
| chr1_+_156552328 | 0.09 |
ENSRNOT00000055401
|
Dlg2
|
discs large MAGUK scaffold protein 2 |
| chr8_-_90984224 | 0.09 |
ENSRNOT00000044931
|
Lca5
|
LCA5, lebercilin |
| chr10_+_42933077 | 0.09 |
ENSRNOT00000003361
|
Mfap3
|
microfibrillar-associated protein 3 |
| chr4_-_130659697 | 0.09 |
ENSRNOT00000072374
|
LOC100363436
|
rCG56280-like |
| chr1_+_87563975 | 0.09 |
ENSRNOT00000088772
|
Zfp30
|
zinc finger protein 30 |
| chr11_+_34865532 | 0.09 |
ENSRNOT00000050342
|
Dyrk1a
|
dual specificity tyrosine phosphorylation regulated kinase 1A |
| chr1_-_7335936 | 0.08 |
ENSRNOT00000020453
|
Ltv1
|
LTV1 ribosome biogenesis factor |
| chr5_+_116420690 | 0.08 |
ENSRNOT00000087089
|
Nfia
|
nuclear factor I/A |
| chr6_-_110669910 | 0.08 |
ENSRNOT00000014635
|
Angel1
|
angel homolog 1 |
| chr3_+_48096954 | 0.08 |
ENSRNOT00000068238
ENSRNOT00000064344 ENSRNOT00000044638 |
Slc4a10
|
solute carrier family 4 member 10 |
| chrX_-_17964888 | 0.08 |
ENSRNOT00000052281
|
LOC100909485
|
spindlin-2-like |
| chr6_+_99883959 | 0.08 |
ENSRNOT00000010588
|
Fntb
|
farnesyltransferase, CAAX box, beta |
| chr1_-_32141036 | 0.08 |
ENSRNOT00000078039
|
Slc12a7
|
solute carrier family 12 member 7 |
| chr3_-_66335869 | 0.08 |
ENSRNOT00000079781
ENSRNOT00000043238 ENSRNOT00000073412 ENSRNOT00000057878 ENSRNOT00000079212 |
Cerkl
|
ceramide kinase-like |
| chrX_+_43535737 | 0.08 |
ENSRNOT00000087073
|
LOC102557137
|
E3 ubiquitin-protein ligase RNF168-like |
| chr4_+_163112301 | 0.08 |
ENSRNOT00000087113
|
Clec12a
|
C-type lectin domain family 12, member A |
| chrX_+_908044 | 0.08 |
ENSRNOT00000072087
|
Zfp300
|
zinc finger protein 300 |
| chr6_-_21112734 | 0.08 |
ENSRNOT00000079819
|
Rasgrp3
|
RAS guanyl releasing protein 3 |
| chr15_-_43542939 | 0.08 |
ENSRNOT00000012996
|
Dpysl2
|
dihydropyrimidinase-like 2 |
| chr1_-_91663467 | 0.08 |
ENSRNOT00000033396
|
Faap24
|
Fanconi anemia core complex associated protein 24 |
| chr4_+_29639154 | 0.08 |
ENSRNOT00000073687
|
Casd1
|
CAS1 domain containing 1 |
| chr8_-_21453190 | 0.08 |
ENSRNOT00000078192
|
Zfp26
|
zinc finger protein 26 |
| chr1_+_171797516 | 0.08 |
ENSRNOT00000088110
|
Ppfibp2
|
PPFIA binding protein 2 |
| chr17_-_89923423 | 0.08 |
ENSRNOT00000076964
|
Acbd5
|
acyl-CoA binding domain containing 5 |
| chr7_+_48867664 | 0.08 |
ENSRNOT00000005862
|
Ppfia2
|
PTPRF interacting protein alpha 2 |
| chr7_+_78188912 | 0.08 |
ENSRNOT00000043079
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr7_-_97994586 | 0.08 |
ENSRNOT00000077616
|
Wdyhv1
|
WDYHV motif containing 1 |
| chrX_-_84768463 | 0.08 |
ENSRNOT00000088570
|
Chm
|
CHM, Rab escort protein 1 |
| chr4_+_138269142 | 0.08 |
ENSRNOT00000007788
|
Cntn4
|
contactin 4 |
| chr16_-_19583386 | 0.08 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chr17_-_56068125 | 0.07 |
ENSRNOT00000022462
|
Mtpap
|
mitochondrial poly(A) polymerase |
| chr4_-_150485781 | 0.07 |
ENSRNOT00000008763
|
Zfp248
|
zinc finger protein 248 |
| chr2_-_140387505 | 0.07 |
ENSRNOT00000047635
|
Elf2
|
E74-like factor 2 |
| chr5_+_4373626 | 0.07 |
ENSRNOT00000064946
|
Eya1
|
EYA transcriptional coactivator and phosphatase 1 |
| chr2_+_250995517 | 0.07 |
ENSRNOT00000018905
|
Odf2l
|
outer dense fiber of sperm tails 2-like |
| chr14_-_28856214 | 0.07 |
ENSRNOT00000044659
|
Adgrl3
|
adhesion G protein-coupled receptor L3 |
| chr15_+_39638510 | 0.07 |
ENSRNOT00000037800
|
Rcbtb1
|
RCC1 and BTB domain containing protein 1 |
| chr1_-_175657485 | 0.07 |
ENSRNOT00000024561
|
rnf141
|
ring finger protein 141 |
| chr16_-_49820235 | 0.07 |
ENSRNOT00000029628
|
Sorbs2
|
sorbin and SH3 domain containing 2 |
| chr6_-_98606109 | 0.07 |
ENSRNOT00000006860
|
Wdr89
|
WD repeat domain 89 |
| chr20_-_45126062 | 0.07 |
ENSRNOT00000000720
|
RGD1310495
|
similar to KIAA1919 protein |
| chr18_-_35780519 | 0.07 |
ENSRNOT00000079667
|
Mcc
|
mutated in colorectal cancers |
| chr3_-_71798531 | 0.07 |
ENSRNOT00000088170
|
Calcrl
|
calcitonin receptor like receptor |
| chr7_+_59200918 | 0.07 |
ENSRNOT00000085073
|
Ptprr
|
protein tyrosine phosphatase, receptor type, R |
| chr5_-_33664435 | 0.07 |
ENSRNOT00000009047
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chr7_+_91384187 | 0.07 |
ENSRNOT00000005828
|
Utp23
|
UTP23, small subunit processome component |
| chr14_+_94590765 | 0.07 |
ENSRNOT00000072533
|
LOC690700
|
similar to similar to RIKEN cDNA 1700001E04 |
| chr18_-_41389510 | 0.07 |
ENSRNOT00000005476
ENSRNOT00000005446 |
Sema6a
|
semaphorin 6A |
| chr2_+_206392200 | 0.07 |
ENSRNOT00000026631
|
Rsbn1
|
round spermatid basic protein 1 |
| chr5_+_122019301 | 0.07 |
ENSRNOT00000068158
|
Pde4b
|
phosphodiesterase 4B |
| chr10_+_42614713 | 0.07 |
ENSRNOT00000081136
ENSRNOT00000073148 |
Gria1
|
glutamate ionotropic receptor AMPA type subunit 1 |
| chrX_-_23530003 | 0.07 |
ENSRNOT00000050819
|
Shroom2
|
shroom family member 2 |
| chr5_+_122647281 | 0.07 |
ENSRNOT00000066041
|
Mier1
|
mesoderm induction early response 1, transcriptional regulator |
| chr18_+_17403407 | 0.07 |
ENSRNOT00000045150
|
RGD1562608
|
similar to KIAA1328 protein |
| chrX_+_123770337 | 0.06 |
ENSRNOT00000092554
|
Nkap
|
NFKB activating protein |
| chr7_+_12840938 | 0.06 |
ENSRNOT00000039232
|
Polrmt
|
RNA polymerase mitochondrial |
| chr18_+_30840868 | 0.06 |
ENSRNOT00000027026
|
Pcdhga5
|
protocadherin gamma subfamily A, 5 |
| chr16_+_6806170 | 0.06 |
ENSRNOT00000084713
|
Sfmbt1
|
Scm-like with four mbt domains 1 |
| chr2_+_12516702 | 0.06 |
ENSRNOT00000050072
|
Tmem161b
|
transmembrane protein 161B |
| chr2_+_248398917 | 0.06 |
ENSRNOT00000045855
|
Gbp1
|
guanylate binding protein 1 |
| chr14_+_48768537 | 0.06 |
ENSRNOT00000082599
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_+_62703779 | 0.06 |
ENSRNOT00000014832
|
Nup205
|
nucleoporin 205 |
| chr9_-_9142339 | 0.06 |
ENSRNOT00000046852
|
MGC116197
|
similar to RIKEN cDNA 1700001E04 |
| chr9_+_1313209 | 0.06 |
ENSRNOT00000083387
|
Tbc1d5
|
TBC1 domain family, member 5 |
| chr11_+_57486775 | 0.06 |
ENSRNOT00000086393
|
LOC103693563
|
mycophenolic acid acyl-glucuronide esterase, mitochondrial |
| chr2_+_118910587 | 0.06 |
ENSRNOT00000065518
|
Zfp639
|
zinc finger protein 639 |
| chr15_+_55043608 | 0.06 |
ENSRNOT00000091000
|
Rcbtb2
|
RCC1 and BTB domain containing protein 2 |
| chr20_+_4043741 | 0.06 |
ENSRNOT00000000525
|
RT1-Bb
|
RT1 class II, locus Bb |
| chr5_-_22799349 | 0.06 |
ENSRNOT00000076204
|
Asph
|
aspartate-beta-hydroxylase |
| chr9_+_12346117 | 0.06 |
ENSRNOT00000074599
|
AABR07066677.1
|
|
| chr6_+_111076351 | 0.06 |
ENSRNOT00000089644
|
Tmem63c
|
transmembrane protein 63c |
| chr14_+_17123360 | 0.06 |
ENSRNOT00000003070
|
Nup54
|
nucleoporin 54 |
| chr4_+_31229913 | 0.05 |
ENSRNOT00000077134
ENSRNOT00000087897 |
Samd9l
|
sterile alpha motif domain containing 9-like |
| chr12_+_23514981 | 0.05 |
ENSRNOT00000001937
|
Prkrip1
|
Prkr interacting protein 1 (IL11 inducible) |
| chr10_-_74769637 | 0.05 |
ENSRNOT00000008889
|
LOC102555183
|
zinc finger protein OZF-like |
| chr7_+_107413691 | 0.05 |
ENSRNOT00000007587
|
Phf20l1
|
PHD finger protein 20-like protein 1 |
| chr10_+_65351245 | 0.05 |
ENSRNOT00000045107
|
Fam222b
|
family with sequence similarity 222, member B |
| chr20_-_46871946 | 0.05 |
ENSRNOT00000031047
|
Armc2
|
armadillo repeat containing 2 |
| chr5_+_134679713 | 0.05 |
ENSRNOT00000067566
|
Mob3c
|
MOB kinase activator 3C |
| chr2_-_257425242 | 0.05 |
ENSRNOT00000017381
|
Dnajb4
|
DnaJ heat shock protein family (Hsp40) member B4 |
| chr8_+_117737117 | 0.05 |
ENSRNOT00000028039
|
Pfkfb4
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 |
| chr19_+_755460 | 0.05 |
ENSRNOT00000076560
|
Dync1li2
|
dynein, cytoplasmic 1 light intermediate chain 2 |
| chr2_-_113345577 | 0.05 |
ENSRNOT00000034096
|
Fndc3b
|
fibronectin type III domain containing 3B |
| chr2_+_27003783 | 0.05 |
ENSRNOT00000061507
|
Poc5
|
POC5 centriolar protein |
| chrX_+_128550178 | 0.05 |
ENSRNOT00000082970
|
Stag2
|
stromal antigen 2 |
| chr4_-_163227242 | 0.05 |
ENSRNOT00000091552
ENSRNOT00000077793 |
Clec7a
|
C-type lectin domain family 7, member A |
| chr14_-_72025137 | 0.05 |
ENSRNOT00000080604
|
C1qtnf7
|
C1q and tumor necrosis factor related protein 7 |
| chr5_-_12199283 | 0.05 |
ENSRNOT00000007769
|
Pcmtd1
|
protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 1 |
| chr10_+_34277993 | 0.05 |
ENSRNOT00000055872
ENSRNOT00000003343 |
Ifi47
|
interferon gamma inducible protein 47 |
| chr1_-_112436794 | 0.05 |
ENSRNOT00000019971
|
Gabrg3
|
gamma-aminobutyric acid type A receptor gamma 3 subunit |
| chr2_+_138194136 | 0.05 |
ENSRNOT00000014928
|
RGD1307595
|
similar to RIKEN cDNA 1700018B24 |
| chr1_+_61786900 | 0.05 |
ENSRNOT00000090287
|
AABR07001905.1
|
|
| chr5_+_120568242 | 0.05 |
ENSRNOT00000050597
|
Lepr
|
leptin receptor |
| chr11_+_72829238 | 0.05 |
ENSRNOT00000071060
|
LOC100910732
|
serine/threonine-protein kinase PAK 2-like |
| chr4_+_106323089 | 0.05 |
ENSRNOT00000091402
|
AABR07061134.1
|
|
| chr2_+_197715761 | 0.05 |
ENSRNOT00000083069
|
Golph3l
|
golgi phosphoprotein 3-like |
| chr7_-_130151414 | 0.05 |
ENSRNOT00000010467
|
Plxnb2
|
plexin B2 |
| chr1_+_30863217 | 0.04 |
ENSRNOT00000015421
|
Rnf146
|
ring finger protein 146 |
| chr15_+_28695912 | 0.04 |
ENSRNOT00000081830
|
Tox4
|
TOX high mobility group box family member 4 |
| chr4_-_135069970 | 0.04 |
ENSRNOT00000008221
|
Cntn3
|
contactin 3 |
| chr13_+_82552550 | 0.04 |
ENSRNOT00000003817
ENSRNOT00000076722 ENSRNOT00000076607 |
Slc19a2
|
solute carrier family 19 member 2 |
| chr3_-_38090526 | 0.04 |
ENSRNOT00000059430
|
Cacnb4
|
calcium voltage-gated channel auxiliary subunit beta 4 |
| chr2_-_240866689 | 0.04 |
ENSRNOT00000036838
|
Nfkb1
|
nuclear factor kappa B subunit 1 |
| chr5_-_120083904 | 0.04 |
ENSRNOT00000064542
|
Jak1
|
Janus kinase 1 |
| chr1_-_126211439 | 0.04 |
ENSRNOT00000014988
|
Tjp1
|
tight junction protein 1 |
| chr8_-_21384131 | 0.04 |
ENSRNOT00000071010
|
Zfp560
|
zinc finger protein 560 |
| chr6_+_100337226 | 0.04 |
ENSRNOT00000011220
|
Fut8
|
fucosyltransferase 8 |
| chr20_+_3149114 | 0.04 |
ENSRNOT00000084770
|
RT1-N2
|
RT1 class Ib, locus N2 |
| chr1_+_199351628 | 0.04 |
ENSRNOT00000078578
|
Bckdk
|
branched chain ketoacid dehydrogenase kinase |
| chr10_+_61685241 | 0.04 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr1_+_99561523 | 0.04 |
ENSRNOT00000038229
|
Ceacam18
|
carcinoembryonic antigen-related cell adhesion molecule 18 |
| chr18_+_44737154 | 0.04 |
ENSRNOT00000021972
|
Tnfaip8
|
TNF alpha induced protein 8 |
| chr13_-_79890134 | 0.04 |
ENSRNOT00000083043
|
RGD1309106
|
similar to hypothetical protein |
| chr18_+_30398113 | 0.04 |
ENSRNOT00000027206
|
Pcdhb5
|
protocadherin beta 5 |
| chr8_+_4440876 | 0.04 |
ENSRNOT00000049325
ENSRNOT00000076529 ENSRNOT00000076748 |
Pdgfd
|
platelet derived growth factor D |
| chr10_-_13845001 | 0.04 |
ENSRNOT00000014269
|
Mlst8
|
MTOR associated protein, LST8 homolog |
| chr14_+_48740190 | 0.04 |
ENSRNOT00000031638
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr4_-_108717309 | 0.04 |
ENSRNOT00000085062
|
AABR07061178.1
|
|
| chr5_+_140870140 | 0.04 |
ENSRNOT00000074347
|
Hpcal4
|
hippocalcin-like 4 |
| chr12_+_10063117 | 0.04 |
ENSRNOT00000044038
|
Usp12
|
ubiquitin specific peptidase 12 |
| chrX_+_44907521 | 0.04 |
ENSRNOT00000004901
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pou2f1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0031959 | mineralocorticoid receptor signaling pathway(GO:0031959) |
| 0.1 | 0.2 | GO:0002835 | negative regulation of T cell mediated cytotoxicity(GO:0001915) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) cytotoxic T cell degranulation(GO:0043316) positive regulation of activation-induced cell death of T cells(GO:0070237) insulin catabolic process(GO:1901143) |
| 0.1 | 0.2 | GO:0046338 | phosphatidylethanolamine catabolic process(GO:0046338) |
| 0.0 | 0.2 | GO:1990962 | carbohydrate export(GO:0033231) daunorubicin transport(GO:0043215) response to borneol(GO:1905230) cellular response to borneol(GO:1905231) response to codeine(GO:1905233) response to quercetin(GO:1905235) drug transport across blood-brain barrier(GO:1990962) establishment of blood-retinal barrier(GO:1990963) |
| 0.0 | 0.1 | GO:0048319 | axial mesoderm morphogenesis(GO:0048319) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:0021836 | chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.2 | GO:0051919 | positive regulation of fibrinolysis(GO:0051919) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.4 | GO:0060693 | regulation of branching involved in salivary gland morphogenesis(GO:0060693) |
| 0.0 | 0.0 | GO:0071226 | cellular response to molecule of fungal origin(GO:0071226) |
| 0.0 | 0.1 | GO:0022605 | oogenesis stage(GO:0022605) |
| 0.0 | 0.1 | GO:0002457 | T cell antigen processing and presentation(GO:0002457) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.1 | GO:0046959 | habituation(GO:0046959) |
| 0.0 | 0.1 | GO:1903385 | ERBB2 signaling pathway(GO:0038128) regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.3 | GO:0021707 | cerebellar granular layer formation(GO:0021684) cerebellar granule cell differentiation(GO:0021707) |
| 0.0 | 0.2 | GO:0043619 | regulation of transcription from RNA polymerase II promoter in response to oxidative stress(GO:0043619) |
| 0.0 | 0.1 | GO:0070995 | NADPH oxidation(GO:0070995) |
| 0.0 | 0.0 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.0 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.1 | GO:0002344 | peripheral B cell selection(GO:0002343) B cell affinity maturation(GO:0002344) |
| 0.0 | 0.1 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0099566 | regulation of postsynaptic cytosolic calcium ion concentration(GO:0099566) |
| 0.0 | 0.1 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.0 | GO:0035696 | monocyte extravasation(GO:0035696) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.0 | GO:1903999 | negative regulation of eating behavior(GO:1903999) regulation of locomotor rhythm(GO:1904059) |
| 0.0 | 0.2 | GO:0051764 | actin filament network formation(GO:0051639) actin crosslink formation(GO:0051764) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.0 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.2 | GO:0006003 | fructose 2,6-bisphosphate metabolic process(GO:0006003) |
| 0.0 | 0.0 | GO:0046368 | GDP-L-fucose metabolic process(GO:0046368) |
| 0.0 | 0.1 | GO:1900041 | negative regulation of interleukin-2 secretion(GO:1900041) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.2 | GO:0014731 | spectrin(GO:0008091) spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.1 | GO:0097441 | basilar dendrite(GO:0097441) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.0 | GO:0042642 | actomyosin, myosin complex part(GO:0042642) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:2001070 | starch binding(GO:2001070) |
| 0.0 | 0.2 | GO:0051377 | mannose-ethanolamine phosphotransferase activity(GO:0051377) |
| 0.0 | 0.2 | GO:0001729 | ceramide kinase activity(GO:0001729) |
| 0.0 | 0.1 | GO:0005302 | L-tyrosine transmembrane transporter activity(GO:0005302) |
| 0.0 | 0.2 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.1 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0002134 | UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0102390 | mycophenolic acid acyl-glucuronide esterase activity(GO:0102390) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0019002 | GMP binding(GO:0019002) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.0 | GO:0015563 | thiamine transmembrane transporter activity(GO:0015234) uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.2 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.1 | GO:0008318 | protein prenyltransferase activity(GO:0008318) |
| 0.0 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.3 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.3 | GO:0005234 | extracellular-glutamate-gated ion channel activity(GO:0005234) |
| 0.0 | 0.0 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID HIF1A PATHWAY | Hypoxic and oxygen homeostasis regulation of HIF-1-alpha |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PE | Genes involved in Acyl chain remodelling of PE |