Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
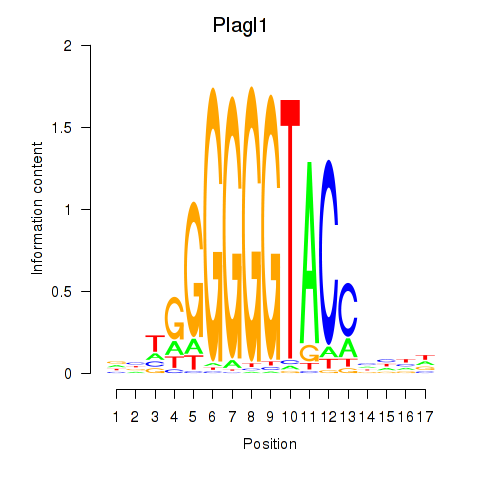
Results for Plagl1
Z-value: 0.30
Transcription factors associated with Plagl1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Plagl1
|
ENSRNOG00000025587 | PLAG1 like zinc finger 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Plagl1 | rn6_v1_chr1_+_7252349_7252349 | 0.18 | 7.7e-01 | Click! |
Activity profile of Plagl1 motif
Sorted Z-values of Plagl1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_11018160 | 0.10 |
ENSRNOT00000092061
|
Aes
|
amino-terminal enhancer of split |
| chr8_-_132027006 | 0.09 |
ENSRNOT00000050963
|
LOC367195
|
similar to 60S RIBOSOMAL PROTEIN L7 |
| chr18_+_32594958 | 0.09 |
ENSRNOT00000018511
|
Spry4
|
sprouty RTK signaling antagonist 4 |
| chr17_-_13393243 | 0.07 |
ENSRNOT00000018252
|
Gadd45g
|
growth arrest and DNA-damage-inducible, gamma |
| chr13_-_50509916 | 0.07 |
ENSRNOT00000076747
|
Ren
|
renin |
| chr1_-_88826302 | 0.07 |
ENSRNOT00000028275
|
Lrfn3
|
leucine rich repeat and fibronectin type III domain containing 3 |
| chr1_-_101236065 | 0.06 |
ENSRNOT00000066834
|
Cd37
|
CD37 molecule |
| chr16_+_41079444 | 0.06 |
ENSRNOT00000015623
|
Neil3
|
nei-like DNA glycosylase 3 |
| chr5_-_36683356 | 0.05 |
ENSRNOT00000009043
|
Pou3f2
|
POU class 3 homeobox 2 |
| chr18_+_29191731 | 0.05 |
ENSRNOT00000068085
ENSRNOT00000025495 |
Cystm1
|
cysteine-rich transmembrane module containing 1 |
| chr5_-_72287669 | 0.05 |
ENSRNOT00000022255
|
Klf4
|
Kruppel like factor 4 |
| chr12_-_21760292 | 0.05 |
ENSRNOT00000059592
|
LOC102550456
|
TSC22 domain family protein 4-like |
| chr13_+_52887649 | 0.05 |
ENSRNOT00000048033
|
Ascl5
|
achaete-scute family bHLH transcription factor 5 |
| chr1_-_101085884 | 0.05 |
ENSRNOT00000085352
|
Rcn3
|
reticulocalbin 3 |
| chr7_-_144936803 | 0.04 |
ENSRNOT00000055279
|
Gpr84
|
G protein-coupled receptor 84 |
| chr19_+_56220755 | 0.04 |
ENSRNOT00000023452
|
Tubb3
|
tubulin, beta 3 class III |
| chr3_+_160852164 | 0.04 |
ENSRNOT00000019127
|
Rbpjl
|
recombination signal binding protein for immunoglobulin kappa J region-like |
| chr7_-_12918173 | 0.04 |
ENSRNOT00000011010
|
Tpgs1
|
tubulin polyglutamylase complex subunit 1 |
| chr2_-_188660179 | 0.04 |
ENSRNOT00000027935
|
Efna4
|
ephrin A4 |
| chr14_+_80248140 | 0.04 |
ENSRNOT00000010852
|
Htra3
|
HtrA serine peptidase 3 |
| chr20_+_44060731 | 0.04 |
ENSRNOT00000000737
|
Lama4
|
laminin subunit alpha 4 |
| chr12_-_9990284 | 0.04 |
ENSRNOT00000001264
|
Rasl11a
|
RAS-like family 11 member A |
| chr20_-_7219548 | 0.04 |
ENSRNOT00000033907
|
Rps10
|
ribosomal protein S10 |
| chr2_-_219558675 | 0.04 |
ENSRNOT00000020149
|
Rtcd1
|
RNA terminal phosphate cyclase domain 1 |
| chr1_-_220806433 | 0.04 |
ENSRNOT00000038138
|
Drap1
|
Dr1 associated protein 1 |
| chr18_-_12640716 | 0.04 |
ENSRNOT00000020697
|
Klhl14
|
kelch-like family member 14 |
| chr10_-_66020682 | 0.04 |
ENSRNOT00000011019
|
Fam58b
|
family with sequence similarity 58, member B |
| chr10_-_14092289 | 0.04 |
ENSRNOT00000019624
|
Ndufb10
|
NADH:ubiquinone oxidoreductase subunit B10 |
| chr13_+_88644520 | 0.03 |
ENSRNOT00000003979
|
Spata46
|
spermatogenesis associated 46 |
| chr10_-_55783489 | 0.03 |
ENSRNOT00000010415
|
Alox15b
|
arachidonate 15-lipoxygenase, type B |
| chr2_+_228544418 | 0.03 |
ENSRNOT00000013030
|
Tram1l1
|
translocation associated membrane protein 1-like 1 |
| chr7_-_107385528 | 0.03 |
ENSRNOT00000093352
|
Tmem71
|
transmembrane protein 71 |
| chr12_-_6703979 | 0.03 |
ENSRNOT00000061246
|
Tex26
|
testis expressed 26 |
| chrX_+_68774699 | 0.03 |
ENSRNOT00000081662
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr10_-_45534570 | 0.03 |
ENSRNOT00000058362
|
Gjc2
|
gap junction protein, gamma 2 |
| chr10_-_88266210 | 0.03 |
ENSRNOT00000090702
ENSRNOT00000020603 |
Hap1
|
huntingtin-associated protein 1 |
| chr9_+_92618352 | 0.03 |
ENSRNOT00000034603
|
Sp140
|
SP140 nuclear body protein |
| chr20_+_6356423 | 0.03 |
ENSRNOT00000000628
|
Cdkn1a
|
cyclin-dependent kinase inhibitor 1A |
| chr1_+_175445088 | 0.03 |
ENSRNOT00000036718
|
Adm
|
adrenomedullin |
| chr13_+_90184569 | 0.03 |
ENSRNOT00000082393
|
Slamf1
|
signaling lymphocytic activation molecule family member 1 |
| chr9_+_94228960 | 0.03 |
ENSRNOT00000083137
ENSRNOT00000080897 |
Akp3
|
alkaline phosphatase 3, intestine, not Mn requiring |
| chr16_+_6970342 | 0.03 |
ENSRNOT00000061294
ENSRNOT00000048344 |
Itih4
|
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr3_+_100788806 | 0.02 |
ENSRNOT00000083289
ENSRNOT00000090445 |
Bdnf
|
brain-derived neurotrophic factor |
| chr9_-_9702306 | 0.02 |
ENSRNOT00000082341
|
Trip10
|
thyroid hormone receptor interactor 10 |
| chr12_-_36555694 | 0.02 |
ENSRNOT00000001292
|
Aacs
|
acetoacetyl-CoA synthetase |
| chr15_+_104095179 | 0.02 |
ENSRNOT00000093487
|
Cldn10
|
claudin 10 |
| chr1_-_222189604 | 0.02 |
ENSRNOT00000028704
|
Kcnk4
|
potassium two pore domain channel subfamily K member 4 |
| chr1_+_101859346 | 0.02 |
ENSRNOT00000028627
|
Kdelr1
|
KDEL endoplasmic reticulum protein retention receptor 1 |
| chr13_+_80517536 | 0.02 |
ENSRNOT00000004386
|
Myoc
|
myocilin |
| chr5_+_133221139 | 0.02 |
ENSRNOT00000047522
|
Trabd2b
|
TraB domain containing 2B |
| chr1_+_7252349 | 0.02 |
ENSRNOT00000030329
|
Plagl1
|
PLAG1 like zinc finger 1 |
| chr9_+_113699170 | 0.02 |
ENSRNOT00000017915
|
Twsg1
|
twisted gastrulation BMP signaling modulator 1 |
| chr5_+_14415606 | 0.02 |
ENSRNOT00000089273
|
Rgs20
|
regulator of G-protein signaling 20 |
| chr4_+_157230769 | 0.02 |
ENSRNOT00000091464
ENSRNOT00000017472 |
Phb2
|
prohibitin 2 |
| chr5_-_170679315 | 0.02 |
ENSRNOT00000071459
|
Ajap1
|
adherens junctions associated protein 1 |
| chr4_+_42202838 | 0.02 |
ENSRNOT00000082453
|
Mdfic
|
MyoD family inhibitor domain containing |
| chr9_-_15306465 | 0.02 |
ENSRNOT00000019404
|
Frs3
|
fibroblast growth factor receptor substrate 3 |
| chrX_-_11164915 | 0.02 |
ENSRNOT00000005138
|
Atp6ap2
|
ATPase H+ transporting accessory protein 2 |
| chr10_+_59533480 | 0.02 |
ENSRNOT00000087723
|
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr2_+_30664441 | 0.02 |
ENSRNOT00000061166
|
Ak6
|
adenylate kinase 6 |
| chr1_-_227175096 | 0.02 |
ENSRNOT00000054811
|
AABR07006259.1
|
|
| chr16_+_51748970 | 0.02 |
ENSRNOT00000059182
|
Adam26a
|
a disintegrin and metallopeptidase domain 26A |
| chr14_-_86796378 | 0.02 |
ENSRNOT00000092021
|
Myo1g
|
myosin IG |
| chr4_-_85192834 | 0.01 |
ENSRNOT00000043752
|
Ggct
|
gamma-glutamyl cyclotransferase |
| chr10_-_103816287 | 0.01 |
ENSRNOT00000004477
|
Grin2c
|
glutamate ionotropic receptor NMDA type subunit 2C |
| chr4_+_98027554 | 0.01 |
ENSRNOT00000009503
|
Serbp1
|
Serpine1 mRNA binding protein 1 |
| chr10_+_59529785 | 0.01 |
ENSRNOT00000064840
ENSRNOT00000065181 |
Atp2a3
|
ATPase sarcoplasmic/endoplasmic reticulum Ca2+ transporting 3 |
| chr17_-_70045865 | 0.01 |
ENSRNOT00000031504
|
Calml5
|
calmodulin-like 5 |
| chr20_+_11114164 | 0.01 |
ENSRNOT00000001602
|
Agpat3
|
1-acylglycerol-3-phosphate O-acyltransferase 3 |
| chr4_-_157230647 | 0.01 |
ENSRNOT00000017334
|
Emg1
|
EMG1 N1-specific pseudouridine methyltransferase |
| chr3_-_10694649 | 0.01 |
ENSRNOT00000037742
|
Gpr107
|
G protein-coupled receptor 107 |
| chr1_-_185143272 | 0.01 |
ENSRNOT00000027752
|
Nucb2
|
nucleobindin 2 |
| chr10_+_56187020 | 0.01 |
ENSRNOT00000046490
|
Tp53
|
tumor protein p53 |
| chr3_-_161212188 | 0.01 |
ENSRNOT00000065751
|
Wfdc3
|
WAP four-disulfide core domain 3 |
| chr15_+_47455690 | 0.01 |
ENSRNOT00000075276
|
LOC100910401
|
serine protease 55-like |
| chr20_+_14578605 | 0.01 |
ENSRNOT00000041165
|
Rtdr1
|
rhabdoid tumor deletion region gene 1 |
| chr1_+_206900617 | 0.01 |
ENSRNOT00000054897
|
Dock1
|
dedicator of cyto-kinesis 1 |
| chr10_-_5253336 | 0.01 |
ENSRNOT00000085310
|
Ciita
|
class II, major histocompatibility complex, transactivator |
| chr3_-_83306781 | 0.01 |
ENSRNOT00000014088
|
Ttc17
|
tetratricopeptide repeat domain 17 |
| chr13_+_48745860 | 0.01 |
ENSRNOT00000010242
|
Slc45a3
|
solute carrier family 45, member 3 |
| chr3_+_15560712 | 0.01 |
ENSRNOT00000010218
|
Ptgs1
|
prostaglandin-endoperoxide synthase 1 |
| chr4_+_21862313 | 0.01 |
ENSRNOT00000007948
|
Dmtf1
|
cyclin D binding myb-like transcription factor 1 |
| chr13_-_85443976 | 0.01 |
ENSRNOT00000005213
|
Uck2
|
uridine-cytidine kinase 2 |
| chr13_+_47572219 | 0.01 |
ENSRNOT00000088449
ENSRNOT00000087664 ENSRNOT00000005853 |
Pigr
|
polymeric immunoglobulin receptor |
| chr20_+_4156858 | 0.01 |
ENSRNOT00000039553
|
Btnl3
|
butyrophilin-like 3 |
| chr7_+_117409576 | 0.01 |
ENSRNOT00000017067
|
Cyc1
|
cytochrome c-1 |
| chr12_+_22026075 | 0.01 |
ENSRNOT00000029041
|
LOC100910838
|
neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adapter 1-like |
| chr20_-_18060661 | 0.01 |
ENSRNOT00000070925
|
AABR07044711.1
|
|
| chr1_+_226687258 | 0.01 |
ENSRNOT00000079679
|
Vwce
|
von Willebrand factor C and EGF domains |
| chr1_-_205630073 | 0.01 |
ENSRNOT00000037064
|
Tex36
|
testis expressed 36 |
| chr7_-_18612118 | 0.01 |
ENSRNOT00000078122
ENSRNOT00000010197 |
Rab11b
|
RAB11B, member RAS oncogene family |
| chr14_-_37763712 | 0.01 |
ENSRNOT00000030610
|
Zar1
|
zygote arrest 1 |
| chr1_-_221269017 | 0.01 |
ENSRNOT00000028363
ENSRNOT00000084396 |
Dpf2
|
double PHD fingers 2 |
| chr6_-_23404368 | 0.01 |
ENSRNOT00000036374
|
Fam179a
|
family with sequence similarity 179, member A |
| chr7_-_115946425 | 0.00 |
ENSRNOT00000000199
|
Jrk
|
jerky homolog (mouse) |
| chr8_-_96088364 | 0.00 |
ENSRNOT00000086161
ENSRNOT00000056818 |
Snx14
|
sorting nexin 14 |
| chr13_-_36101411 | 0.00 |
ENSRNOT00000074471
|
Tmem37
|
transmembrane protein 37 |
| chr12_+_23463157 | 0.00 |
ENSRNOT00000044841
|
Cux1
|
cut-like homeobox 1 |
| chr1_+_212578450 | 0.00 |
ENSRNOT00000064501
|
Paox
|
polyamine oxidase |
| chr1_+_170212817 | 0.00 |
ENSRNOT00000040672
|
RGD1561034
|
similar to hypothetical protein MGC34805 |
| chr10_+_104377748 | 0.00 |
ENSRNOT00000080714
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr15_-_56970365 | 0.00 |
ENSRNOT00000047192
|
Lrch1
|
leucine rich repeats and calponin homology domain containing 1 |
| chr1_+_31700953 | 0.00 |
ENSRNOT00000020251
|
Exoc3
|
exocyst complex component 3 |
| chr12_+_23151180 | 0.00 |
ENSRNOT00000059486
|
Cux1
|
cut-like homeobox 1 |
| chr7_+_11095468 | 0.00 |
ENSRNOT00000061219
|
Celf5
|
CUGBP, Elav-like family member 5 |
| chr6_-_33088582 | 0.00 |
ENSRNOT00000083144
|
Tdrd15
|
tudor domain containing 15 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Plagl1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0002003 | angiotensin maturation(GO:0002003) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0001010 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) |