Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
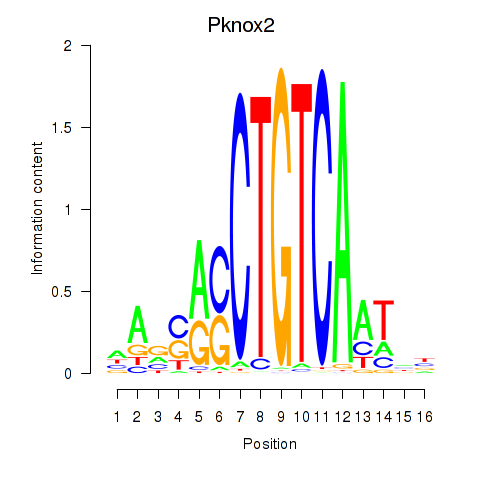
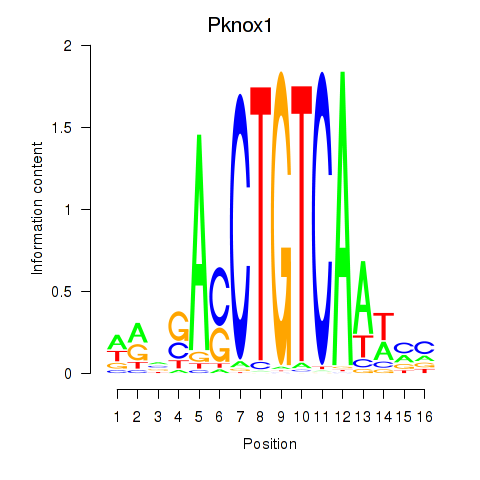
Results for Pknox2_Pknox1
Z-value: 0.20
Transcription factors associated with Pknox2_Pknox1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pknox2
|
ENSRNOG00000028856 | PBX/knotted 1 homeobox 2 |
|
Pknox1
|
ENSRNOG00000001184 | PBX/knotted 1 homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pknox2 | rn6_v1_chr8_-_39551700_39551700 | 0.32 | 6.0e-01 | Click! |
| Pknox1 | rn6_v1_chr20_+_10334308_10334308 | 0.10 | 8.7e-01 | Click! |
Activity profile of Pknox2_Pknox1 motif
Sorted Z-values of Pknox2_Pknox1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_-_140618405 | 0.12 |
ENSRNOT00000017736
|
Setd7
|
SET domain containing (lysine methyltransferase) 7 |
| chr12_+_660011 | 0.07 |
ENSRNOT00000040830
|
Pds5b
|
PDS5 cohesin associated factor B |
| chr10_+_59360765 | 0.07 |
ENSRNOT00000036278
|
Zzef1
|
zinc finger ZZ-type and EF-hand domain containing 1 |
| chr2_+_198797159 | 0.06 |
ENSRNOT00000056225
|
Ankrd35
|
ankyrin repeat domain 35 |
| chr1_-_201981229 | 0.06 |
ENSRNOT00000077246
ENSRNOT00000035531 |
Ikzf5
|
IKAROS family zinc finger 5 |
| chr2_+_155555840 | 0.06 |
ENSRNOT00000080951
|
AABR07010944.1
|
|
| chr18_+_44468784 | 0.05 |
ENSRNOT00000031812
|
Dmxl1
|
Dmx-like 1 |
| chr10_+_90731865 | 0.05 |
ENSRNOT00000064429
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr3_-_125213607 | 0.05 |
ENSRNOT00000078070
|
Gpcpd1
|
glycerophosphocholine phosphodiesterase 1 |
| chr17_-_43614844 | 0.05 |
ENSRNOT00000023054
|
Hist1h1a
|
histone cluster 1 H1 family member a |
| chr4_-_59809321 | 0.04 |
ENSRNOT00000017536
|
Plxna4
|
plexin A4 |
| chr20_+_46428124 | 0.04 |
ENSRNOT00000000327
|
Foxo3
|
forkhead box O3 |
| chr13_+_97838361 | 0.04 |
ENSRNOT00000003641
|
Cnst
|
consortin, connexin sorting protein |
| chr8_-_33017854 | 0.04 |
ENSRNOT00000011386
|
Barx2
|
BARX homeobox 2 |
| chr4_+_127164453 | 0.03 |
ENSRNOT00000017889
|
Kbtbd8
|
kelch repeat and BTB domain containing 8 |
| chr12_-_17186679 | 0.03 |
ENSRNOT00000001730
|
Uncx
|
UNC homeobox |
| chr7_-_120967490 | 0.03 |
ENSRNOT00000046399
|
Sun2
|
Sad1 and UNC84 domain containing 2 |
| chr3_+_8958090 | 0.03 |
ENSRNOT00000086789
ENSRNOT00000064557 |
Dolpp1
|
dolichyldiphosphatase 1 |
| chr5_+_151573092 | 0.03 |
ENSRNOT00000011049
|
Slc9a1
|
solute carrier family 9 member A1 |
| chr7_-_94563001 | 0.03 |
ENSRNOT00000051139
ENSRNOT00000005561 |
Enpp2
|
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr17_-_18421861 | 0.03 |
ENSRNOT00000060500
ENSRNOT00000036560 |
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr7_+_66595742 | 0.03 |
ENSRNOT00000031191
|
Usp15
|
ubiquitin specific peptidase 15 |
| chr1_-_265573117 | 0.02 |
ENSRNOT00000044195
ENSRNOT00000055915 |
LOC100911951
|
Kv channel-interacting protein 2-like |
| chr2_+_3400977 | 0.02 |
ENSRNOT00000093593
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chr9_+_94178221 | 0.02 |
ENSRNOT00000033487
|
Alppl2
|
alkaline phosphatase, placental-like 2 |
| chr2_-_219628997 | 0.02 |
ENSRNOT00000064484
|
Trmt13
|
tRNA methyltransferase 13 homolog |
| chr8_+_94122728 | 0.02 |
ENSRNOT00000014783
|
Dopey1
|
dopey family member 1 |
| chr10_+_4945911 | 0.02 |
ENSRNOT00000003420
|
Prm1
|
protamine 1 |
| chr9_-_45206427 | 0.02 |
ENSRNOT00000042227
|
Aff3
|
AF4/FMR2 family, member 3 |
| chr2_-_43999107 | 0.02 |
ENSRNOT00000065473
|
LOC100912399
|
mitogen-activated protein kinase kinase kinase 1-like |
| chr9_+_17163354 | 0.02 |
ENSRNOT00000026049
|
Polh
|
DNA polymerase eta |
| chr4_+_181315444 | 0.02 |
ENSRNOT00000044147
|
Ppfibp1
|
PPFIA binding protein 1 |
| chr6_+_52265930 | 0.02 |
ENSRNOT00000087513
ENSRNOT00000067849 |
Sypl1
|
synaptophysin-like 1 |
| chr5_+_124442293 | 0.02 |
ENSRNOT00000041922
|
RGD1564074
|
similar to novel protein |
| chr1_+_197999037 | 0.02 |
ENSRNOT00000091065
|
Apobr
|
apolipoprotein B receptor |
| chr12_-_17358617 | 0.02 |
ENSRNOT00000001733
|
Gpr146
|
G protein-coupled receptor 146 |
| chr10_+_90731148 | 0.02 |
ENSRNOT00000093604
|
Adam11
|
ADAM metallopeptidase domain 11 |
| chr17_-_10208360 | 0.02 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr6_+_58468155 | 0.02 |
ENSRNOT00000091263
|
Etv1
|
ets variant 1 |
| chr10_+_36715565 | 0.02 |
ENSRNOT00000005048
|
Clk4
|
CDC-like kinase 4 |
| chr4_-_62526724 | 0.01 |
ENSRNOT00000038950
|
RGD1565367
|
similar to Solute carrier family 23, member 2 (Sodium-dependent vitamin C transporter 2) |
| chr14_-_81254637 | 0.01 |
ENSRNOT00000058166
|
Htt
|
huntingtin |
| chr9_-_17163170 | 0.01 |
ENSRNOT00000025921
|
Xpo5
|
exportin 5 |
| chr12_+_39253409 | 0.01 |
ENSRNOT00000001774
|
Camkk2
|
calcium/calmodulin-dependent protein kinase kinase 2 |
| chr1_+_65564173 | 0.01 |
ENSRNOT00000038860
|
Zbtb45
|
zinc finger and BTB domain containing 45 |
| chr14_+_83897117 | 0.01 |
ENSRNOT00000026569
|
Morc2
|
MORC family CW-type zinc finger 2 |
| chr8_-_76239995 | 0.01 |
ENSRNOT00000071321
|
LOC100912027
|
60S ribosomal protein L27-like |
| chr3_+_8802852 | 0.01 |
ENSRNOT00000033934
|
Lrrc8a
|
leucine rich repeat containing 8 family, member A |
| chrX_+_29562165 | 0.01 |
ENSRNOT00000006074
|
Ofd1
|
OFD1, centriole and centriolar satellite protein |
| chr14_+_83412968 | 0.01 |
ENSRNOT00000024993
|
Eif4enif1
|
eukaryotic translation initiation factor 4E nuclear import factor 1 |
| chr8_-_59077690 | 0.01 |
ENSRNOT00000066197
|
Dmxl2
|
Dmx-like 2 |
| chr1_+_153589471 | 0.01 |
ENSRNOT00000023205
|
Fzd4
|
frizzled class receptor 4 |
| chr11_-_31238026 | 0.01 |
ENSRNOT00000032366
|
RGD1562726
|
similar to Putative protein C21orf62 homolog |
| chr13_+_113373578 | 0.01 |
ENSRNOT00000009900
|
Plxna2
|
plexin A2 |
| chr20_+_46667121 | 0.01 |
ENSRNOT00000089611
|
Sesn1
|
sestrin 1 |
| chr15_+_103344476 | 0.01 |
ENSRNOT00000013329
ENSRNOT00000077047 ENSRNOT00000078103 |
Gpr180
|
G protein-coupled receptor 180 |
| chr13_-_48284408 | 0.01 |
ENSRNOT00000085967
|
Srgap2
|
SLIT-ROBO Rho GTPase activating protein 2 |
| chr2_+_179952227 | 0.01 |
ENSRNOT00000015081
|
Pdgfc
|
platelet derived growth factor C |
| chr1_+_214454090 | 0.01 |
ENSRNOT00000049448
|
Tspan4
|
tetraspanin 4 |
| chr6_+_64808238 | 0.01 |
ENSRNOT00000093195
|
Nrcam
|
neuronal cell adhesion molecule |
| chr3_+_143003796 | 0.01 |
ENSRNOT00000006287
|
Gzf1
|
GDNF-inducible zinc finger protein 1 |
| chr3_+_175144495 | 0.01 |
ENSRNOT00000082601
ENSRNOT00000088026 |
Cdh4
|
cadherin 4 |
| chr6_+_113898420 | 0.01 |
ENSRNOT00000064872
|
Nrxn3
|
neurexin 3 |
| chr6_-_55001464 | 0.01 |
ENSRNOT00000006618
|
Ahr
|
aryl hydrocarbon receptor |
| chr4_-_183374195 | 0.01 |
ENSRNOT00000073927
ENSRNOT00000074740 ENSRNOT00000070907 |
Caprin2
|
caprin family member 2 |
| chr7_+_108613739 | 0.01 |
ENSRNOT00000007564
|
Phf20l1
|
PHD finger protein 20-like 1 |
| chr14_+_79261092 | 0.01 |
ENSRNOT00000029191
|
LOC680039
|
hypothetical protein LOC680039 |
| chr2_-_188727670 | 0.01 |
ENSRNOT00000074920
|
Lenep
|
lens epithelial protein |
| chr13_-_97838228 | 0.01 |
ENSRNOT00000003618
|
Tfb2m
|
transcription factor B2, mitochondrial |
| chr3_+_122544788 | 0.01 |
ENSRNOT00000063828
|
Tgm3
|
transglutaminase 3 |
| chr2_+_140471690 | 0.01 |
ENSRNOT00000017379
|
Naa15
|
N(alpha)-acetyltransferase 15, NatA auxiliary subunit |
| chr9_-_74048244 | 0.01 |
ENSRNOT00000018293
|
Lancl1
|
LanC like 1 |
| chr20_-_5212624 | 0.01 |
ENSRNOT00000074261
|
LOC103689996
|
antigen peptide transporter 2 |
| chr2_+_218951451 | 0.00 |
ENSRNOT00000019190
|
Extl2
|
exostosin-like glycosyltransferase 2 |
| chr10_-_59360661 | 0.00 |
ENSRNOT00000036942
|
Cyb5d2
|
cytochrome b5 domain containing 2 |
| chr6_-_137733026 | 0.00 |
ENSRNOT00000019213
|
Jag2
|
jagged 2 |
| chr3_-_8979889 | 0.00 |
ENSRNOT00000065128
|
Crat
|
carnitine O-acetyltransferase |
| chr1_-_198320075 | 0.00 |
ENSRNOT00000081291
|
Taok2
|
TAO kinase 2 |
| chr13_-_95250235 | 0.00 |
ENSRNOT00000085648
|
Akt3
|
AKT serine/threonine kinase 3 |
| chr1_-_57555048 | 0.00 |
ENSRNOT00000066370
|
Prdm9
|
PR/SET domain 9 |
| chr15_-_88670349 | 0.00 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr10_+_88245532 | 0.00 |
ENSRNOT00000019863
|
Gast
|
gastrin |
| chr15_+_51222954 | 0.00 |
ENSRNOT00000064428
ENSRNOT00000088928 |
Entpd4
|
ectonucleoside triphosphate diphosphohydrolase 4 |
| chr1_+_93242050 | 0.00 |
ENSRNOT00000013741
|
RGD1562402
|
similar to 60S ribosomal protein L27a |
| chr10_-_46969406 | 0.00 |
ENSRNOT00000006753
|
Flii
|
FLII, actin remodeling protein |
| chr7_-_11266630 | 0.00 |
ENSRNOT00000027926
|
Cactin
|
cactin, spliceosome C complex subunit |
| chr11_+_84745904 | 0.00 |
ENSRNOT00000002617
|
Klhl6
|
kelch-like family member 6 |
| chr18_+_51523758 | 0.00 |
ENSRNOT00000078518
|
Gramd3
|
GRAM domain containing 3 |
| chr14_-_84820415 | 0.00 |
ENSRNOT00000057501
|
Mtmr3
|
myotubularin related protein 3 |
| chr20_+_3995544 | 0.00 |
ENSRNOT00000000527
|
Tap2
|
transporter 2, ATP binding cassette subfamily B member |
| chr6_+_132090218 | 0.00 |
ENSRNOT00000083208
|
Ccnk
|
cyclin K |
| chr12_+_41316764 | 0.00 |
ENSRNOT00000090867
|
Oas3
|
2'-5'-oligoadenylate synthetase 3 |
| chr14_-_105055421 | 0.00 |
ENSRNOT00000008274
|
Lgalsl
|
galectin-like |
| chr6_+_132090633 | 0.00 |
ENSRNOT00000057171
|
Ccnk
|
cyclin K |
| chr7_-_60743328 | 0.00 |
ENSRNOT00000066767
|
Mdm2
|
MDM2 proto-oncogene |
| chr7_+_12146642 | 0.00 |
ENSRNOT00000085899
|
Tcf3
|
transcription factor 3 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pknox2_Pknox1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0018026 | peptidyl-lysine monomethylation(GO:0018026) |
| 0.0 | 0.0 | GO:0030473 | nucleokinesis involved in cell motility in cerebral cortex radial glia guided migration(GO:0021817) nuclear migration along microtubule(GO:0030473) nuclear matrix anchoring at nuclear membrane(GO:0090292) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:2001070 | starch binding(GO:2001070) |