Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
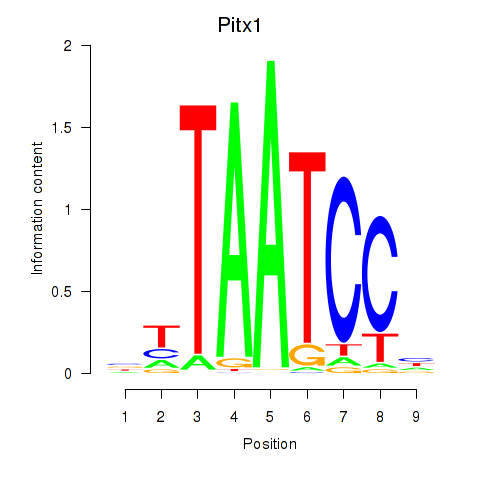
Results for Pitx1
Z-value: 0.76
Transcription factors associated with Pitx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pitx1
|
ENSRNOG00000011423 | paired-like homeodomain 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pitx1 | rn6_v1_chr17_+_8878270_8878270 | 0.63 | 2.5e-01 | Click! |
Activity profile of Pitx1 motif
Sorted Z-values of Pitx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_48438259 | 0.89 |
ENSRNOT00000059813
|
Mfrp
|
membrane frizzled-related protein |
| chr8_+_48437918 | 0.67 |
ENSRNOT00000085578
|
Mfrp
|
membrane frizzled-related protein |
| chr2_-_48501436 | 0.48 |
ENSRNOT00000017305
|
Isl1
|
ISL LIM homeobox 1 |
| chr1_-_166943592 | 0.45 |
ENSRNOT00000026962
|
Folr1
|
folate receptor 1 |
| chr10_-_63176463 | 0.31 |
ENSRNOT00000004717
|
Slc6a4
|
solute carrier family 6 member 4 |
| chr1_-_24191908 | 0.30 |
ENSRNOT00000061157
|
Sgk1
|
serum/glucocorticoid regulated kinase 1 |
| chr8_-_108109801 | 0.27 |
ENSRNOT00000034277
|
Sox14
|
SRY box 14 |
| chr2_+_145174876 | 0.27 |
ENSRNOT00000040631
|
Mab21l1
|
mab-21 like 1 |
| chr9_-_121972055 | 0.24 |
ENSRNOT00000089735
|
AABR07068851.1
|
clusterin-like protein 1 |
| chr10_-_87407634 | 0.23 |
ENSRNOT00000016657
|
Krt23
|
keratin 23 |
| chr4_-_30276372 | 0.22 |
ENSRNOT00000011823
|
Pon1
|
paraoxonase 1 |
| chr12_+_25264400 | 0.20 |
ENSRNOT00000087656
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr8_+_55050284 | 0.20 |
ENSRNOT00000013242
|
Pih1d2
|
PIH1 domain containing 2 |
| chr4_+_28972434 | 0.19 |
ENSRNOT00000014219
|
Gngt1
|
G protein subunit gamma transducin 1 |
| chr7_-_120490061 | 0.19 |
ENSRNOT00000016247
|
Slc16a8
|
solute carrier family 16 member 8 |
| chr4_+_30313102 | 0.18 |
ENSRNOT00000012657
|
Asb4
|
ankyrin repeat and SOCS box-containing 4 |
| chr10_-_87195075 | 0.17 |
ENSRNOT00000014851
|
Krt24
|
keratin 24 |
| chr5_+_135997052 | 0.16 |
ENSRNOT00000024921
|
Tctex1d4
|
Tctex1 domain containing 4 |
| chr5_+_138300107 | 0.16 |
ENSRNOT00000047151
|
Cldn19
|
claudin 19 |
| chr15_+_44441856 | 0.16 |
ENSRNOT00000018006
|
Gnrh1
|
gonadotropin releasing hormone 1 |
| chrX_+_105402867 | 0.15 |
ENSRNOT00000057222
|
Rpl36a
|
ribosomal protein L36a |
| chr8_+_94686938 | 0.15 |
ENSRNOT00000013285
|
Ripply2
|
ripply transcriptional repressor 2 |
| chr8_+_117953444 | 0.14 |
ENSRNOT00000028141
|
Cdc25a
|
cell division cycle 25A |
| chr10_+_105630810 | 0.14 |
ENSRNOT00000064904
|
Prcd
|
photoreceptor disc component |
| chr3_+_112242270 | 0.14 |
ENSRNOT00000080533
ENSRNOT00000082876 |
Capn3
|
calpain 3 |
| chr5_+_29538380 | 0.14 |
ENSRNOT00000010845
|
Calb1
|
calbindin 1 |
| chr10_-_48038647 | 0.14 |
ENSRNOT00000078448
|
Prpsap2
|
phosphoribosyl pyrophosphate synthetase-associated protein 2 |
| chr5_-_135025084 | 0.13 |
ENSRNOT00000018766
|
Tspan1
|
tetraspanin 1 |
| chr3_+_91870208 | 0.13 |
ENSRNOT00000040249
|
Rpl21
|
ribosomal protein L21 |
| chr18_-_59819113 | 0.13 |
ENSRNOT00000065939
|
RGD1562699
|
RGD1562699 |
| chr4_+_136512201 | 0.13 |
ENSRNOT00000051645
|
Cntn6
|
contactin 6 |
| chr10_+_104368247 | 0.12 |
ENSRNOT00000006519
|
Llgl2
|
LLGL2, scribble cell polarity complex component |
| chr12_-_22126858 | 0.12 |
ENSRNOT00000084480
|
Sap25
|
Sin3A-associated protein 25 |
| chr5_+_21830882 | 0.11 |
ENSRNOT00000008901
|
Chd7
|
chromodomain helicase DNA binding protein 7 |
| chr19_-_46167950 | 0.11 |
ENSRNOT00000071024
|
AABR07043892.1
|
|
| chr12_+_25264192 | 0.11 |
ENSRNOT00000079392
|
Gtf2ird1
|
GTF2I repeat domain containing 1 |
| chr2_+_225645568 | 0.11 |
ENSRNOT00000017878
|
Abca4
|
ATP binding cassette subfamily A member 4 |
| chr11_-_43099412 | 0.10 |
ENSRNOT00000002281
|
Gabrr3
|
gamma-aminobutyric acid type A receptor rho3 subunit |
| chr10_+_69737328 | 0.10 |
ENSRNOT00000055999
ENSRNOT00000076773 |
Tmem132e
|
transmembrane protein 132E |
| chr8_-_117353672 | 0.10 |
ENSRNOT00000027262
|
Ndufaf3
|
NADH:ubiquinone oxidoreductase complex assembly factor 3 |
| chr4_+_94696965 | 0.10 |
ENSRNOT00000064696
|
Grid2
|
glutamate ionotropic receptor delta type subunit 2 |
| chr1_+_89008117 | 0.10 |
ENSRNOT00000028401
|
Hspb6
|
heat shock protein family B (small) member 6 |
| chr3_-_44086006 | 0.10 |
ENSRNOT00000034449
ENSRNOT00000082604 |
Ermn
|
ermin |
| chr7_+_60015998 | 0.10 |
ENSRNOT00000007309
|
Best3
|
bestrophin 3 |
| chr4_+_151298548 | 0.09 |
ENSRNOT00000010746
|
Cacna2d4
|
calcium voltage-gated channel auxiliary subunit alpha2delta 4 |
| chr4_-_51199570 | 0.09 |
ENSRNOT00000010788
|
Slc13a1
|
solute carrier family 13 member 1 |
| chr13_+_85818427 | 0.09 |
ENSRNOT00000077227
ENSRNOT00000006117 |
Rxrg
|
retinoid X receptor gamma |
| chr3_-_113423149 | 0.08 |
ENSRNOT00000021008
|
Serinc4
|
serine incorporator 4 |
| chr7_+_143874734 | 0.08 |
ENSRNOT00000093277
|
Gm9918
|
predicted gene 9918 |
| chr2_+_95008311 | 0.08 |
ENSRNOT00000077270
|
Tpd52
|
tumor protein D52 |
| chr9_+_81816872 | 0.08 |
ENSRNOT00000041407
|
Plcd4
|
phospholipase C, delta 4 |
| chr5_-_115387377 | 0.08 |
ENSRNOT00000036030
ENSRNOT00000077492 |
RGD1560146
|
similar to hypothetical protein MGC34837 |
| chr16_+_23317953 | 0.08 |
ENSRNOT00000075287
|
AABR07024972.1
|
|
| chr5_+_133864798 | 0.08 |
ENSRNOT00000091977
|
Tal1
|
TAL bHLH transcription factor 1, erythroid differentiation factor |
| chr7_+_94795214 | 0.08 |
ENSRNOT00000005722
|
Deptor
|
DEP domain containing MTOR-interacting protein |
| chr18_+_36713869 | 0.08 |
ENSRNOT00000025465
|
Pou4f3
|
POU class 4 homeobox 3 |
| chr8_-_68525911 | 0.08 |
ENSRNOT00000080588
ENSRNOT00000011109 |
Iqch
|
IQ motif containing H |
| chr2_+_241909832 | 0.07 |
ENSRNOT00000047975
|
Ppp3ca
|
protein phosphatase 3 catalytic subunit alpha |
| chr3_+_113318563 | 0.07 |
ENSRNOT00000089230
|
Ckmt1
|
creatine kinase, mitochondrial 1 |
| chr5_+_128190708 | 0.07 |
ENSRNOT00000012492
|
Orc1
|
origin recognition complex, subunit 1 |
| chr10_-_56000225 | 0.07 |
ENSRNOT00000013439
|
Tmem88
|
transmembrane protein 88 |
| chr5_+_98387291 | 0.07 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr18_-_24929091 | 0.07 |
ENSRNOT00000019596
|
Proc
|
protein C |
| chr1_+_107344904 | 0.07 |
ENSRNOT00000082582
|
Gas2
|
growth arrest-specific 2 |
| chr20_-_5806097 | 0.07 |
ENSRNOT00000000611
|
Clps
|
colipase |
| chr7_-_49741540 | 0.07 |
ENSRNOT00000006523
|
Myf6
|
myogenic factor 6 |
| chr20_-_7943575 | 0.07 |
ENSRNOT00000066897
|
Tulp1
|
tubby like protein 1 |
| chr4_+_147832136 | 0.07 |
ENSRNOT00000064603
|
Rho
|
rhodopsin |
| chr7_-_44771458 | 0.06 |
ENSRNOT00000006007
|
Alx1
|
ALX homeobox 1 |
| chr2_+_60966789 | 0.06 |
ENSRNOT00000025490
|
Slc45a2
|
solute carrier family 45, member 2 |
| chr4_+_117962319 | 0.06 |
ENSRNOT00000057441
|
Tgfa
|
transforming growth factor alpha |
| chr14_+_23480462 | 0.06 |
ENSRNOT00000002755
|
Gnrhr
|
gonadotropin releasing hormone receptor |
| chr3_-_91195981 | 0.06 |
ENSRNOT00000056935
|
RGD1309730
|
similar to RIKEN cDNA B230118H07 |
| chr2_+_45104305 | 0.06 |
ENSRNOT00000014559
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr2_-_259382765 | 0.06 |
ENSRNOT00000091407
|
St6galnac3
|
ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 3 |
| chr2_+_24449485 | 0.06 |
ENSRNOT00000073688
|
Tbca
|
tubulin folding cofactor A |
| chr3_-_104018861 | 0.06 |
ENSRNOT00000008387
|
Chrm5
|
cholinergic receptor, muscarinic 5 |
| chr11_+_87220618 | 0.06 |
ENSRNOT00000041975
|
Gsc2
|
goosecoid homeobox 2 |
| chr7_-_117732339 | 0.06 |
ENSRNOT00000092917
ENSRNOT00000020696 |
Foxh1
|
forkhead box H1 |
| chr8_-_107490093 | 0.06 |
ENSRNOT00000046832
|
LOC684466
|
similar to Fas apoptotic inhibitory molecule 1 (rFAIM) |
| chr9_-_93125014 | 0.06 |
ENSRNOT00000023829
|
Htr2b
|
5-hydroxytryptamine receptor 2B |
| chr15_+_3938075 | 0.06 |
ENSRNOT00000065644
|
Camk2g
|
calcium/calmodulin-dependent protein kinase II gamma |
| chr19_-_38120578 | 0.06 |
ENSRNOT00000026873
|
Esrp2
|
epithelial splicing regulatory protein 2 |
| chrX_+_70461718 | 0.06 |
ENSRNOT00000078233
ENSRNOT00000003789 |
Kif4a
|
kinesin family member 4A |
| chr1_+_137799185 | 0.05 |
ENSRNOT00000083590
ENSRNOT00000092778 |
Agbl1
|
ATP/GTP binding protein-like 1 |
| chr16_+_10277775 | 0.05 |
ENSRNOT00000090679
|
Rbp3
|
retinol binding protein 3 |
| chr3_-_50120392 | 0.05 |
ENSRNOT00000006175
|
Fign
|
fidgetin, microtubule severing factor |
| chr4_-_56656980 | 0.05 |
ENSRNOT00000009278
|
Opn1sw
|
opsin 1 (cone pigments), short-wave-sensitive |
| chr4_+_14001761 | 0.05 |
ENSRNOT00000076519
|
Cd36
|
CD36 molecule |
| chrY_-_1305026 | 0.05 |
ENSRNOT00000092901
|
Usp9y
|
ubiquitin specific peptidase 9, Y-linked |
| chr2_-_206222248 | 0.05 |
ENSRNOT00000026075
|
Olfml3
|
olfactomedin-like 3 |
| chr9_+_47185443 | 0.05 |
ENSRNOT00000020194
|
Il18r1
|
interleukin 18 receptor 1 |
| chr3_+_80075991 | 0.05 |
ENSRNOT00000080266
|
Pacsin3
|
protein kinase C and casein kinase substrate in neurons 3 |
| chr4_-_170740274 | 0.05 |
ENSRNOT00000012212
|
Gucy2c
|
guanylate cyclase 2C |
| chr5_+_15043955 | 0.05 |
ENSRNOT00000047093
|
Rp1
|
retinitis pigmentosa 1 |
| chr8_-_68275720 | 0.04 |
ENSRNOT00000079122
|
Map2k5
|
mitogen activated protein kinase kinase 5 |
| chr10_+_34578356 | 0.04 |
ENSRNOT00000047454
|
Olr1389
|
olfactory receptor 1389 |
| chr10_-_85289777 | 0.04 |
ENSRNOT00000055427
ENSRNOT00000014863 |
Gpr179
|
G protein-coupled receptor 179 |
| chr2_+_3662763 | 0.04 |
ENSRNOT00000017828
|
Mctp1
|
multiple C2 and transmembrane domain containing 1 |
| chrM_+_8599 | 0.04 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr3_+_117421604 | 0.04 |
ENSRNOT00000008860
ENSRNOT00000008857 |
Slc12a1
|
solute carrier family 12 member 1 |
| chr2_-_203680083 | 0.04 |
ENSRNOT00000021268
|
Cd2
|
Cd2 molecule |
| chr16_+_49185560 | 0.04 |
ENSRNOT00000014360
|
Helt
|
helt bHLH transcription factor |
| chr8_+_33514042 | 0.04 |
ENSRNOT00000081614
ENSRNOT00000081525 |
Kcnj1
|
potassium voltage-gated channel subfamily J member 1 |
| chr20_+_28989491 | 0.04 |
ENSRNOT00000074524
|
Pla2g12b
|
phospholipase A2, group XIIB |
| chr5_+_48303366 | 0.04 |
ENSRNOT00000009973
|
Gabrr2
|
gamma-aminobutyric acid type A receptor rho 2 subunit |
| chr11_+_43521054 | 0.04 |
ENSRNOT00000041679
|
Olr1551
|
olfactory receptor 1551 |
| chr8_+_42511520 | 0.04 |
ENSRNOT00000042974
|
Olr1259
|
olfactory receptor 1259 |
| chrX_-_15741103 | 0.04 |
ENSRNOT00000066489
|
Cacna1f
|
calcium voltage-gated channel subunit alpha1 F |
| chr6_+_56625650 | 0.04 |
ENSRNOT00000008803
|
Meox2
|
mesenchyme homeobox 2 |
| chr2_+_54191538 | 0.04 |
ENSRNOT00000019524
|
Plcxd3
|
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr19_+_10142496 | 0.04 |
ENSRNOT00000088645
ENSRNOT00000060351 |
Cngb1
|
cyclic nucleotide gated channel beta 1 |
| chr3_+_138715570 | 0.03 |
ENSRNOT00000064723
|
Sec23b
|
Sec23 homolog B, coat complex II component |
| chr4_+_8066737 | 0.03 |
ENSRNOT00000063994
|
Srpk2
|
SRSF protein kinase 2 |
| chr12_+_13097269 | 0.03 |
ENSRNOT00000087094
|
Rac1
|
ras-related C3 botulinum toxin substrate 1 |
| chr1_+_277261681 | 0.03 |
ENSRNOT00000022724
|
Plekhs1
|
pleckstrin homology domain containing S1 |
| chr12_-_39697465 | 0.03 |
ENSRNOT00000078683
|
Vps29
|
VPS29 retromer complex component |
| chr19_-_43215281 | 0.03 |
ENSRNOT00000025052
|
Aars
|
alanyl-tRNA synthetase |
| chr18_+_56544652 | 0.03 |
ENSRNOT00000024171
|
Pde6a
|
phosphodiesterase 6A |
| chrX_-_25628272 | 0.03 |
ENSRNOT00000086414
|
Mid1
|
midline 1 |
| chr1_-_134871167 | 0.03 |
ENSRNOT00000076300
|
Chd2
|
chromodomain helicase DNA binding protein 2 |
| chr16_+_14306804 | 0.03 |
ENSRNOT00000017773
|
Lrit1
|
leucine-rich repeat, Ig-like and transmembrane domains 1 |
| chr5_+_104362971 | 0.03 |
ENSRNOT00000058520
|
Adamtsl1
|
ADAMTS-like 1 |
| chr2_+_46066780 | 0.03 |
ENSRNOT00000073226
|
LOC108350225
|
olfactory receptor 8B8-like |
| chr10_-_55851235 | 0.03 |
ENSRNOT00000010790
|
Gucy2d
|
guanylate cyclase 2D, retinal |
| chrX_+_159158194 | 0.03 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr8_-_55171718 | 0.03 |
ENSRNOT00000080736
|
LOC689959
|
hypothetical protein LOC689959 |
| chr16_+_39353283 | 0.03 |
ENSRNOT00000080125
|
LOC108348202
|
disintegrin and metalloproteinase domain-containing protein 21-like |
| chr1_-_8885967 | 0.03 |
ENSRNOT00000016102
|
Gje1
|
gap junction protein, epsilon 1 |
| chr18_-_74542077 | 0.03 |
ENSRNOT00000078519
|
Slc14a2
|
solute carrier family 14 member 2 |
| chr15_-_34647421 | 0.03 |
ENSRNOT00000072426
|
Mcpt8
|
mast cell protease 8 |
| chr20_+_20378861 | 0.03 |
ENSRNOT00000091044
|
Ank3
|
ankyrin 3 |
| chr10_-_88036040 | 0.03 |
ENSRNOT00000018851
|
Krt13
|
keratin 13 |
| chr1_+_124983452 | 0.03 |
ENSRNOT00000021262
|
Trpm1
|
transient receptor potential cation channel, subfamily M, member 1 |
| chr18_-_37096132 | 0.02 |
ENSRNOT00000041188
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chrX_-_156589907 | 0.02 |
ENSRNOT00000083147
|
Opn1mw
|
opsin 1 (cone pigments), medium-wave-sensitive |
| chrX_-_10218583 | 0.02 |
ENSRNOT00000013382
|
Nyx
|
nyctalopin |
| chr4_+_8066907 | 0.02 |
ENSRNOT00000080811
|
Srpk2
|
SRSF protein kinase 2 |
| chr14_+_19319299 | 0.02 |
ENSRNOT00000086542
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr2_-_250805445 | 0.02 |
ENSRNOT00000055362
|
Clca4l
|
chloride channel calcium activated 4-like |
| chr3_+_113423693 | 0.02 |
ENSRNOT00000021091
|
Hypk
|
Huntingtin interacting protein K |
| chr2_+_128461224 | 0.02 |
ENSRNOT00000018872
|
Jade1
|
jade family PHD finger 1 |
| chr16_+_2670618 | 0.02 |
ENSRNOT00000030102
|
Il17rd
|
interleukin 17 receptor D |
| chr8_-_114617466 | 0.01 |
ENSRNOT00000082992
ENSRNOT00000038160 |
Col6a5
|
collagen type VI alpha 5 chain |
| chr19_-_43215077 | 0.01 |
ENSRNOT00000082151
|
Aars
|
alanyl-tRNA synthetase |
| chr18_+_71395830 | 0.01 |
ENSRNOT00000024831
|
Smad7
|
SMAD family member 7 |
| chr20_-_5123073 | 0.01 |
ENSRNOT00000001126
|
Apom
|
apolipoprotein M |
| chr10_-_55560422 | 0.01 |
ENSRNOT00000006883
|
Rangrf
|
RAN guanine nucleotide release factor |
| chr3_-_147849875 | 0.01 |
ENSRNOT00000081153
|
Nrsn2
|
neurensin 2 |
| chr7_-_70476340 | 0.01 |
ENSRNOT00000006800
|
Arhgef25
|
Rho guanine nucleotide exchange factor 25 |
| chr10_+_53818818 | 0.01 |
ENSRNOT00000057260
|
Myh8
|
myosin heavy chain 8 |
| chr18_-_11858744 | 0.01 |
ENSRNOT00000061417
ENSRNOT00000082891 |
Dsc2
|
desmocollin 2 |
| chr3_+_56355431 | 0.01 |
ENSRNOT00000037188
|
Myo3b
|
myosin IIIB |
| chr2_-_45480798 | 0.01 |
ENSRNOT00000066764
|
Snx18
|
sorting nexin 18 |
| chr14_+_108007724 | 0.00 |
ENSRNOT00000014062
|
Xpo1
|
exportin 1 |
| chr14_-_113644779 | 0.00 |
ENSRNOT00000005306
|
Cfap36
|
cilia and flagella associated protein 36 |
| chr4_+_155531906 | 0.00 |
ENSRNOT00000060937
|
Nanog
|
Nanog homeobox |
| chr6_+_23337571 | 0.00 |
ENSRNOT00000011832
|
RGD1304963
|
similar to hypothetical protein MGC38716 |
| chr20_-_3401273 | 0.00 |
ENSRNOT00000089257
ENSRNOT00000078451 ENSRNOT00000001085 |
Nrm
|
nurim (nuclear envelope membrane protein) |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pitx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0071657 | visceral motor neuron differentiation(GO:0021524) sensory system development(GO:0048880) positive regulation of granulocyte colony-stimulating factor production(GO:0071657) positive regulation of macrophage colony-stimulating factor production(GO:1901258) |
| 0.1 | 0.4 | GO:0060974 | cell migration involved in heart formation(GO:0060974) |
| 0.1 | 0.2 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) negative regulation of lipoprotein oxidation(GO:0034443) |
| 0.1 | 0.3 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) |
| 0.0 | 0.1 | GO:1990091 | sodium-dependent self proteolysis(GO:1990091) |
| 0.0 | 0.3 | GO:0014886 | transition between slow and fast fiber(GO:0014886) |
| 0.0 | 1.8 | GO:0042462 | eye photoreceptor cell development(GO:0042462) |
| 0.0 | 0.1 | GO:0006649 | phospholipid transfer to membrane(GO:0006649) |
| 0.0 | 0.1 | GO:0035502 | metanephric part of ureteric bud development(GO:0035502) |
| 0.0 | 0.3 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 0.2 | GO:2000354 | regulation of ovarian follicle development(GO:2000354) |
| 0.0 | 0.1 | GO:0016332 | establishment or maintenance of polarity of embryonic epithelium(GO:0016332) |
| 0.0 | 0.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:1905203 | regulation of connective tissue replacement(GO:1905203) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0035054 | embryonic heart tube anterior/posterior pattern specification(GO:0035054) |
| 0.0 | 0.2 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.1 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.1 | GO:0036302 | ventricular compact myocardium morphogenesis(GO:0003223) atrioventricular canal development(GO:0036302) |
| 0.0 | 0.1 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.2 | GO:2001214 | positive regulation of vasculogenesis(GO:2001214) |
| 0.0 | 0.0 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.0 | GO:0032978 | protein insertion into membrane from inner side(GO:0032978) |
| 0.0 | 0.0 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.0 | 0.2 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.0 | GO:0035655 | interleukin-18-mediated signaling pathway(GO:0035655) cellular response to interleukin-18(GO:0071351) |
| 0.0 | 0.1 | GO:0007207 | adenylate cyclase-inhibiting G-protein coupled acetylcholine receptor signaling pathway(GO:0007197) phospholipase C-activating G-protein coupled acetylcholine receptor signaling pathway(GO:0007207) |
| 0.0 | 0.2 | GO:0050908 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.1 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.0 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:1990005 | granular vesicle(GO:1990005) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0034366 | spherical high-density lipoprotein particle(GO:0034366) |
| 0.0 | 0.4 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.2 | GO:0031466 | Cul5-RING ubiquitin ligase complex(GO:0031466) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
| 0.0 | 0.1 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0051870 | methotrexate binding(GO:0051870) |
| 0.1 | 0.3 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.2 | GO:0046573 | lactonohydrolase activity(GO:0046573) acyl-L-homoserine-lactone lactonohydrolase activity(GO:0102007) |
| 0.0 | 0.3 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.0 | 0.1 | GO:0099510 | calcium ion binding involved in regulation of cytosolic calcium ion concentration(GO:0099510) |
| 0.0 | 0.2 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0070012 | oligopeptidase activity(GO:0070012) |
| 0.0 | 0.1 | GO:0090555 | phosphatidylethanolamine-translocating ATPase activity(GO:0090555) |
| 0.0 | 0.1 | GO:0015154 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.5 | GO:0016922 | ligand-dependent nuclear receptor binding(GO:0016922) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 0.1 | GO:0033192 | calcium-dependent protein serine/threonine phosphatase activity(GO:0004723) calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.1 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.1 | GO:0016716 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, another compound as one donor, and incorporation of one atom of oxygen(GO:0016716) |
| 0.0 | 0.1 | GO:0009881 | photoreceptor activity(GO:0009881) |
| 0.0 | 0.1 | GO:0031432 | titin binding(GO:0031432) |
| 0.0 | 0.0 | GO:0008511 | sodium:potassium:chloride symporter activity(GO:0008511) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |
| 0.0 | 0.1 | GO:0016907 | G-protein coupled acetylcholine receptor activity(GO:0016907) |
| 0.0 | 0.0 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.2 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
| 0.0 | 0.1 | SA G2 AND M PHASES | Cdc25 activates the cdc2/cyclin B complex to induce the G2/M transition. |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.5 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | Genes involved in Synthesis, Secretion, and Inactivation of Glucose-dependent Insulinotropic Polypeptide (GIP) |
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.3 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.1 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |