Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
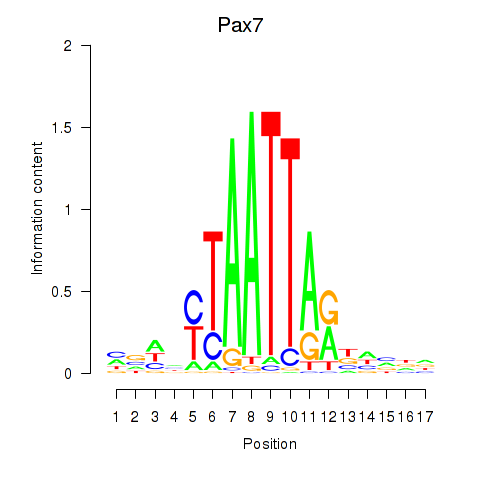
Results for Pax7
Z-value: 0.09
Transcription factors associated with Pax7
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax7
|
ENSRNOG00000018739 | paired box 7 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax7 | rn6_v1_chr5_-_158313426_158313426 | -0.48 | 4.1e-01 | Click! |
Activity profile of Pax7 motif
Sorted Z-values of Pax7 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr2_+_127525285 | 0.03 |
ENSRNOT00000093247
|
Intu
|
inturned planar cell polarity protein |
| chr1_-_190370499 | 0.03 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr18_-_55891710 | 0.03 |
ENSRNOT00000064686
|
Synpo
|
synaptopodin |
| chr5_+_139790395 | 0.03 |
ENSRNOT00000015033
|
Rims3
|
regulating synaptic membrane exocytosis 3 |
| chr12_-_45801842 | 0.03 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr20_-_13994794 | 0.02 |
ENSRNOT00000093466
|
Ggt5
|
gamma-glutamyltransferase 5 |
| chr3_+_14889510 | 0.02 |
ENSRNOT00000080760
|
Dab2ip
|
DAB2 interacting protein |
| chr10_-_88670430 | 0.02 |
ENSRNOT00000025547
|
Hcrt
|
hypocretin neuropeptide precursor |
| chr4_-_18396035 | 0.02 |
ENSRNOT00000034692
|
Sema3a
|
semaphorin 3A |
| chr2_+_104020955 | 0.02 |
ENSRNOT00000045586
|
Mtfr1
|
mitochondrial fission regulator 1 |
| chr10_-_74679858 | 0.02 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr10_-_67401836 | 0.01 |
ENSRNOT00000073071
|
Crlf3
|
cytokine receptor-like factor 3 |
| chr2_-_33025271 | 0.01 |
ENSRNOT00000074941
|
NEWGENE_1310139
|
microtubule associated serine/threonine kinase family member 4 |
| chr19_-_22194740 | 0.01 |
ENSRNOT00000086187
|
Phkb
|
phosphorylase kinase regulatory subunit beta |
| chr8_-_84506328 | 0.01 |
ENSRNOT00000064754
|
Mlip
|
muscular LMNA-interacting protein |
| chr16_+_9023387 | 0.01 |
ENSRNOT00000067761
|
RGD1561145
|
similar to novel protein |
| chr13_-_111581018 | 0.01 |
ENSRNOT00000083072
ENSRNOT00000077981 |
Sertad4
|
SERTA domain containing 4 |
| chr2_-_35870578 | 0.01 |
ENSRNOT00000066069
|
Rnf180
|
ring finger protein 180 |
| chr12_-_35979193 | 0.01 |
ENSRNOT00000071104
|
Tmem132b
|
transmembrane protein 132B |
| chr3_-_90751055 | 0.01 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr7_-_15073052 | 0.01 |
ENSRNOT00000037708
|
Zfp799
|
zinc finger protein 799 |
| chr9_+_71915421 | 0.01 |
ENSRNOT00000020447
|
Pikfyve
|
phosphoinositide kinase, FYVE-type zinc finger containing |
| chr4_+_88694583 | 0.01 |
ENSRNOT00000009202
|
Ppm1k
|
protein phosphatase, Mg2+/Mn2+ dependent, 1K |
| chr4_-_119327822 | 0.01 |
ENSRNOT00000012645
|
Arhgap25
|
Rho GTPase activating protein 25 |
| chr4_-_55011415 | 0.01 |
ENSRNOT00000056996
|
Grm8
|
glutamate metabotropic receptor 8 |
| chr4_-_183424449 | 0.01 |
ENSRNOT00000071930
|
Fam60a
|
family with sequence similarity 60, member A |
| chr7_+_2752680 | 0.01 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr5_+_28485619 | 0.01 |
ENSRNOT00000093341
ENSRNOT00000093129 |
LOC100912373
|
uncharacterized LOC100912373 |
| chr14_-_16903242 | 0.01 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr9_-_30844199 | 0.01 |
ENSRNOT00000017169
|
Col19a1
|
collagen type XIX alpha 1 chain |
| chr10_-_65502936 | 0.01 |
ENSRNOT00000089615
|
Supt6h
|
SPT6 homolog, histone chaperone |
| chr8_-_39551700 | 0.00 |
ENSRNOT00000091894
ENSRNOT00000076025 |
Pknox2
|
PBX/knotted 1 homeobox 2 |
| chr11_-_62067655 | 0.00 |
ENSRNOT00000093382
|
Zbtb20
|
zinc finger and BTB domain containing 20 |
| chr13_-_76049363 | 0.00 |
ENSRNOT00000075865
ENSRNOT00000007455 |
Brinp2
|
BMP/retinoic acid inducible neural specific 2 |
| chr1_+_101554642 | 0.00 |
ENSRNOT00000028474
|
Bcat2
|
branched chain amino acid transaminase 2 |
| chr19_+_39229754 | 0.00 |
ENSRNOT00000050612
|
Vps4a
|
vacuolar protein sorting 4 homolog A |
| chr13_+_91054974 | 0.00 |
ENSRNOT00000091089
|
Crp
|
C-reactive protein |
| chr6_+_101532518 | 0.00 |
ENSRNOT00000075054
|
Gphn
|
gephyrin |
| chr1_+_80141630 | 0.00 |
ENSRNOT00000029552
|
Opa3
|
optic atrophy 3 |
| chr16_+_60925093 | 0.00 |
ENSRNOT00000015813
|
Tnks
|
tankyrase |
| chr10_-_103826448 | 0.00 |
ENSRNOT00000085636
|
Fdxr
|
ferredoxin reductase |
| chr7_-_28711761 | 0.00 |
ENSRNOT00000006249
|
Parpbp
|
PARP1 binding protein |
| chr6_-_108660063 | 0.00 |
ENSRNOT00000006240
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chrX_+_118197217 | 0.00 |
ENSRNOT00000090922
|
Htr2c
|
5-hydroxytryptamine receptor 2C |
| chr18_+_79773608 | 0.00 |
ENSRNOT00000088484
|
Zfp516
|
zinc finger protein 516 |
| chr12_+_47590154 | 0.00 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr1_-_233145924 | 0.00 |
ENSRNOT00000018763
ENSRNOT00000082442 |
Psat1
|
phosphoserine aminotransferase 1 |
| chr2_+_66940057 | 0.00 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr7_-_143967484 | 0.00 |
ENSRNOT00000081758
|
Sp7
|
Sp7 transcription factor |
| chr11_-_61234944 | 0.00 |
ENSRNOT00000059680
|
Cfap44
|
cilia and flagella associated protein 44 |
| chr2_-_122646445 | 0.00 |
ENSRNOT00000080100
|
Dcun1d1
|
defective in cullin neddylation 1 domain containing 1 |
| chr4_+_180291389 | 0.00 |
ENSRNOT00000002465
|
Sspn
|
sarcospan |
| chr9_+_60039297 | 0.00 |
ENSRNOT00000016262
|
Slc39a10
|
solute carrier family 39 member 10 |
| chrX_-_104932508 | 0.00 |
ENSRNOT00000075325
|
Nox1
|
NADPH oxidase 1 |
| chr4_+_156253079 | 0.00 |
ENSRNOT00000013536
|
Clec4d
|
C-type lectin domain family 4, member D |
| chr8_+_22966561 | 0.00 |
ENSRNOT00000016819
|
Swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr7_+_78558701 | 0.00 |
ENSRNOT00000006393
|
Rims2
|
regulating synaptic membrane exocytosis 2 |
| chr6_+_145546595 | 0.00 |
ENSRNOT00000007112
|
Rapgef5
|
Rap guanine nucleotide exchange factor 5 |
| chr8_+_22966792 | 0.00 |
ENSRNOT00000064580
|
Swsap1
|
SWIM-type zinc finger 7 associated protein 1 |
| chr7_-_64887448 | 0.00 |
ENSRNOT00000038147
|
Helb
|
DNA helicase B |
| chr10_-_87153982 | 0.00 |
ENSRNOT00000014414
|
Krt222
|
keratin 222 |
| chr2_+_202200797 | 0.00 |
ENSRNOT00000042263
ENSRNOT00000071938 |
Spag17
|
sperm associated antigen 17 |
| chr18_+_28594892 | 0.00 |
ENSRNOT00000067702
|
Ube2d2
|
ubiquitin-conjugating enzyme E2D 2 |