Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Pax6
Z-value: 0.35
Transcription factors associated with Pax6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax6
|
ENSRNOG00000004410 | paired box 6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax6 | rn6_v1_chr3_+_95715193_95715193 | 0.54 | 3.5e-01 | Click! |
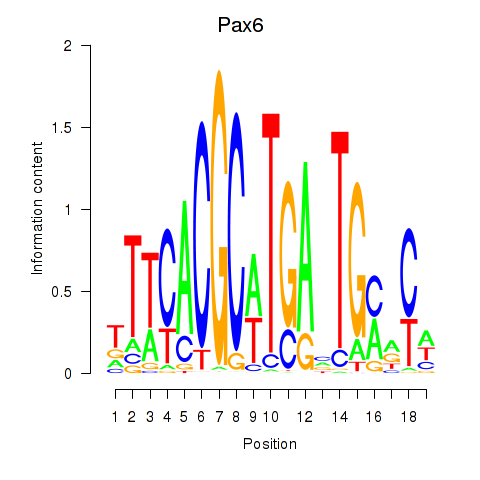
Activity profile of Pax6 motif
Sorted Z-values of Pax6 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_2982603 | 0.13 |
ENSRNOT00000045460
|
Kcnb2
|
potassium voltage-gated channel subfamily B member 2 |
| chr19_+_10563423 | 0.10 |
ENSRNOT00000021037
|
Dok4
|
docking protein 4 |
| chr2_-_41785792 | 0.09 |
ENSRNOT00000015871
|
Rab3c
|
RAB3C, member RAS oncogene family |
| chr1_-_166911694 | 0.08 |
ENSRNOT00000066915
|
Inppl1
|
inositol polyphosphate phosphatase-like 1 |
| chr16_+_35573058 | 0.08 |
ENSRNOT00000059580
|
Galntl6
|
polypeptide N-acetylgalactosaminyltransferase-like 6 |
| chr6_+_106052212 | 0.07 |
ENSRNOT00000010626
|
Sipa1l1
|
signal-induced proliferation-associated 1 like 1 |
| chr5_+_152613255 | 0.07 |
ENSRNOT00000037097
|
Pafah2
|
platelet-activating factor acetylhydrolase 2 |
| chr1_-_87777359 | 0.06 |
ENSRNOT00000074090
|
AABR07002870.1
|
|
| chr8_-_117237229 | 0.06 |
ENSRNOT00000071381
|
Klhdc8b
|
kelch domain containing 8B |
| chr6_-_3254779 | 0.06 |
ENSRNOT00000061975
|
Cdkl4
|
cyclin-dependent kinase-like 4 |
| chr9_-_103207190 | 0.06 |
ENSRNOT00000026010
|
St8sia4
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 |
| chr20_+_3230052 | 0.06 |
ENSRNOT00000078454
|
RT1-T24-3
|
RT1 class I, locus T24, gene 3 |
| chr17_-_44840131 | 0.06 |
ENSRNOT00000083417
|
Hist1h3b
|
histone cluster 1, H3b |
| chr17_-_42031265 | 0.06 |
ENSRNOT00000068021
|
Dcdc2
|
doublecortin domain containing 2 |
| chr3_+_95715193 | 0.06 |
ENSRNOT00000089525
|
Pax6
|
paired box 6 |
| chrX_+_23146085 | 0.06 |
ENSRNOT00000068703
|
Apex2
|
apurinic/apyrimidinic endodeoxyribonuclease 2 |
| chr1_-_87777668 | 0.05 |
ENSRNOT00000037288
|
AABR07002870.1
|
|
| chr11_+_87549059 | 0.05 |
ENSRNOT00000002567
|
Ccdc74a
|
coiled-coil domain containing 74A |
| chr16_-_74864816 | 0.05 |
ENSRNOT00000017164
|
Alg11
|
ALG11, alpha-1,2-mannosyltransferase |
| chr5_-_59553416 | 0.05 |
ENSRNOT00000090490
|
Gne
|
glucosamine (UDP-N-acetyl)-2-epimerase/N-acetylmannosamine kinase |
| chr16_-_19583386 | 0.05 |
ENSRNOT00000090131
|
Zfp617
|
zinc finger protein 617 |
| chrX_+_82143789 | 0.05 |
ENSRNOT00000003724
|
Pou3f4
|
POU class 3 homeobox 4 |
| chr4_+_98457810 | 0.05 |
ENSRNOT00000074175
|
AABR07060872.1
|
|
| chr18_+_30869628 | 0.05 |
ENSRNOT00000060470
|
Pcdhgb4
|
protocadherin gamma subfamily B, 4 |
| chr12_+_37709860 | 0.05 |
ENSRNOT00000086244
|
Mphosph9
|
M-phase phosphoprotein 9 |
| chr6_-_65319527 | 0.05 |
ENSRNOT00000005618
|
Stxbp6
|
syntaxin binding protein 6 |
| chr12_-_13157570 | 0.04 |
ENSRNOT00000001437
ENSRNOT00000067332 ENSRNOT00000092238 |
Daglb
|
diacylglycerol lipase, beta |
| chr1_+_86938138 | 0.04 |
ENSRNOT00000075601
|
Ccer2
|
coiled-coil glutamate-rich protein 2 |
| chr2_+_26240385 | 0.04 |
ENSRNOT00000024292
|
F2rl2
|
coagulation factor II (thrombin) receptor-like 2 |
| chr18_+_55463308 | 0.04 |
ENSRNOT00000073388
|
LOC100910979
|
interferon-inducible GTPase 1-like |
| chr1_-_84812486 | 0.04 |
ENSRNOT00000078369
|
AABR07002779.1
|
|
| chr11_+_43194348 | 0.04 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
| chr1_-_225952516 | 0.04 |
ENSRNOT00000043387
|
Incenp
|
inner centromere protein |
| chr8_-_49280901 | 0.04 |
ENSRNOT00000021390
|
Cd3g
|
CD3g molecule |
| chr6_-_8344897 | 0.04 |
ENSRNOT00000082353
|
Prepl
|
prolyl endopeptidase-like |
| chr1_+_261337594 | 0.04 |
ENSRNOT00000019874
|
Pi4k2a
|
phosphatidylinositol 4-kinase type 2 alpha |
| chr18_+_53088994 | 0.04 |
ENSRNOT00000091921
|
AC104053.2
|
|
| chr12_+_7208850 | 0.04 |
ENSRNOT00000001219
|
Katnal1
|
katanin catalytic subunit A1 like 1 |
| chrX_-_15707436 | 0.04 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr2_+_116032455 | 0.04 |
ENSRNOT00000066495
|
Phc3
|
polyhomeotic homolog 3 |
| chr14_-_33164141 | 0.04 |
ENSRNOT00000002844
|
Rest
|
RE1-silencing transcription factor |
| chrX_+_159158194 | 0.03 |
ENSRNOT00000043820
ENSRNOT00000001169 ENSRNOT00000083502 |
Fhl1
|
four and a half LIM domains 1 |
| chr7_+_42304534 | 0.03 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chr3_+_72238981 | 0.03 |
ENSRNOT00000011006
|
Slc43a1
|
solute carrier family 43 member 1 |
| chr10_-_54967585 | 0.03 |
ENSRNOT00000005255
|
Ntn1
|
netrin 1 |
| chr11_+_76147205 | 0.03 |
ENSRNOT00000071586
|
Fgf12
|
fibroblast growth factor 12 |
| chr19_+_11450760 | 0.03 |
ENSRNOT00000026297
|
Nudt21
|
nudix hydrolase 21 |
| chr3_-_12944494 | 0.03 |
ENSRNOT00000023172
|
Mvb12b
|
multivesicular body subunit 12B |
| chr4_-_56786754 | 0.03 |
ENSRNOT00000050795
|
Kcp
|
kielin/chordin-like protein |
| chr13_-_113872097 | 0.03 |
ENSRNOT00000010799
ENSRNOT00000084320 |
Cr1l
|
complement C3b/C4b receptor 1 like |
| chr2_-_188559882 | 0.03 |
ENSRNOT00000088199
|
Trim46
|
tripartite motif-containing 46 |
| chr4_+_149970567 | 0.03 |
ENSRNOT00000091765
|
Hnrnpf
|
heterogeneous nuclear ribonucleoprotein F |
| chr1_-_154170409 | 0.03 |
ENSRNOT00000089014
|
Hikeshi
|
Hikeshi, heat shock protein nuclear import factor |
| chr2_+_188139730 | 0.03 |
ENSRNOT00000056789
ENSRNOT00000077698 |
Gon4l
|
gon-4 like |
| chr7_-_11699436 | 0.03 |
ENSRNOT00000049221
ENSRNOT00000066037 |
Tmprss9
|
transmembrane protease, serine 9 |
| chr18_+_30808404 | 0.03 |
ENSRNOT00000060475
|
Pcdhga1
|
protocadherin gamma subfamily A, 1 |
| chr13_+_89718066 | 0.03 |
ENSRNOT00000005147
|
Dedd
|
death effector domain-containing |
| chr17_-_2705123 | 0.03 |
ENSRNOT00000024940
|
Olr1652
|
olfactory receptor 1652 |
| chr12_-_36555694 | 0.03 |
ENSRNOT00000001292
|
Aacs
|
acetoacetyl-CoA synthetase |
| chr13_-_93307199 | 0.03 |
ENSRNOT00000065041
|
Rgs7
|
regulator of G-protein signaling 7 |
| chr6_-_102196138 | 0.02 |
ENSRNOT00000014132
|
Tmem229b
|
transmembrane protein 229B |
| chr11_-_84931160 | 0.02 |
ENSRNOT00000071848
|
B3gnt5
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 |
| chr10_-_58771908 | 0.02 |
ENSRNOT00000018808
|
RGD1304728
|
similar to 4933427D14Rik protein |
| chr5_+_134679713 | 0.02 |
ENSRNOT00000067566
|
Mob3c
|
MOB kinase activator 3C |
| chr19_-_11451278 | 0.02 |
ENSRNOT00000026118
|
Ogfod1
|
2-oxoglutarate and iron-dependent oxygenase domain containing 1 |
| chr1_+_226947105 | 0.02 |
ENSRNOT00000028373
|
Prpf19
|
pre-mRNA processing factor 19 |
| chr2_+_78245459 | 0.02 |
ENSRNOT00000089551
|
LOC103689968
|
protein FAM134B |
| chr1_-_216971183 | 0.02 |
ENSRNOT00000077911
|
Mrgpre
|
MAS related GPR family member E |
| chr14_+_86029335 | 0.02 |
ENSRNOT00000017375
|
Dbnl
|
drebrin-like |
| chr7_-_139907640 | 0.02 |
ENSRNOT00000045473
|
Zfp641
|
zinc finger protein 641 |
| chr1_+_1771710 | 0.02 |
ENSRNOT00000080138
ENSRNOT00000073528 |
Nup43
|
nucleoporin 43 |
| chrX_-_77295426 | 0.02 |
ENSRNOT00000090833
|
Taf9b
|
TATA-box binding protein associated factor 9b |
| chr18_+_30880020 | 0.02 |
ENSRNOT00000060468
|
Pcdhgb5
|
protocadherin gamma subfamily B, 5 |
| chr18_+_402295 | 0.02 |
ENSRNOT00000033618
|
Fundc2
|
FUN14 domain containing 2 |
| chr8_-_71533069 | 0.02 |
ENSRNOT00000021863
|
Trip4
|
thyroid hormone receptor interactor 4 |
| chr3_-_52849907 | 0.02 |
ENSRNOT00000041096
|
Scn7a
|
sodium voltage-gated channel alpha subunit 7 |
| chr3_+_128828331 | 0.02 |
ENSRNOT00000045393
|
Plcb4
|
phospholipase C, beta 4 |
| chr10_+_97710905 | 0.02 |
ENSRNOT00000000263
|
Amz2
|
archaelysin family metallopeptidase 2 |
| chr3_+_121632043 | 0.02 |
ENSRNOT00000024882
|
Polr1b
|
RNA polymerase I subunit B |
| chr11_-_84090259 | 0.02 |
ENSRNOT00000002321
|
Eif2b5
|
eukaryotic translation initiation factor 2B subunit 5 epsilon |
| chr4_-_155690869 | 0.02 |
ENSRNOT00000012216
|
C3ar1
|
complement C3a receptor 1 |
| chr2_+_187322416 | 0.02 |
ENSRNOT00000025183
|
Crabp2
|
cellular retinoic acid binding protein 2 |
| chr10_+_46940965 | 0.02 |
ENSRNOT00000005366
|
Llgl1
|
LLGL1, scribble cell polarity complex component |
| chr12_-_13998172 | 0.02 |
ENSRNOT00000001476
|
Wipi2
|
WD repeat domain, phosphoinositide interacting 2 |
| chr5_+_103479767 | 0.02 |
ENSRNOT00000008999
|
Sh3gl2
|
SH3 domain-containing GRB2-like 2 |
| chr8_+_48699769 | 0.02 |
ENSRNOT00000044613
|
Hyou1
|
hypoxia up-regulated 1 |
| chr20_+_4369178 | 0.01 |
ENSRNOT00000088079
|
Agpat1
|
1-acylglycerol-3-phosphate O-acyltransferase 1 |
| chr6_+_128750795 | 0.01 |
ENSRNOT00000005781
|
Glrx5
|
glutaredoxin 5 |
| chr5_+_64892802 | 0.01 |
ENSRNOT00000059869
|
Rnf20
|
ring finger protein 20 |
| chr3_-_64364101 | 0.01 |
ENSRNOT00000089065
|
Zfp385b
|
zinc finger protein 385B |
| chr12_+_38377034 | 0.01 |
ENSRNOT00000088002
|
Clip1
|
CAP-GLY domain containing linker protein 1 |
| chr9_+_52687868 | 0.01 |
ENSRNOT00000083864
|
Wdr75
|
WD repeat domain 75 |
| chr6_+_26546924 | 0.01 |
ENSRNOT00000007763
|
Eif2b4
|
eukaryotic translation initiation factor 2B subunit delta |
| chr8_-_50126413 | 0.01 |
ENSRNOT00000072712
ENSRNOT00000022583 |
Cep164
|
centrosomal protein 164 |
| chr18_+_55666027 | 0.01 |
ENSRNOT00000045950
|
RGD1305184
|
similar to CDNA sequence BC023105 |
| chr13_-_50514151 | 0.01 |
ENSRNOT00000003951
|
Ren
|
renin |
| chr3_-_12467662 | 0.01 |
ENSRNOT00000051893
|
Zbtb34
|
zinc finger and BTB domain containing 34 |
| chr16_+_20704807 | 0.01 |
ENSRNOT00000027241
|
Klhl26
|
kelch-like family member 26 |
| chr4_-_66955732 | 0.01 |
ENSRNOT00000084282
|
Kdm7a
|
lysine (K)-specific demethylase 7A |
| chr13_+_93684437 | 0.01 |
ENSRNOT00000005005
|
Kmo
|
kynurenine 3-monooxygenase |
| chr17_+_49610548 | 0.01 |
ENSRNOT00000017955
|
Yae1d1
|
Yae1 domain containing 1 |
| chr1_-_31528083 | 0.01 |
ENSRNOT00000061041
|
Rpl26-ps2
|
ribosomal protein L26, pseudogene 2 |
| chr1_+_65851060 | 0.01 |
ENSRNOT00000036880
|
Zscan18
|
zinc finger and SCAN domain containing 18 |
| chr5_+_153260930 | 0.01 |
ENSRNOT00000023289
ENSRNOT00000082184 |
Rsrp1
|
arginine and serine rich protein 1 |
| chr10_+_58995556 | 0.01 |
ENSRNOT00000045100
|
Ggt6
|
gamma-glutamyl transferase 6 |
| chr5_-_119564846 | 0.01 |
ENSRNOT00000012977
|
Cyp2j4
|
cytochrome P450, family 2, subfamily j, polypeptide 4 |
| chr1_+_214215992 | 0.01 |
ENSRNOT00000051755
ENSRNOT00000023376 |
Phrf1
|
PHD and ring finger domains 1 |
| chr11_-_79703736 | 0.01 |
ENSRNOT00000044279
|
Lpp
|
LIM domain containing preferred translocation partner in lipoma |
| chr7_+_80750725 | 0.01 |
ENSRNOT00000079962
|
Oxr1
|
oxidation resistance 1 |
| chr15_+_18941431 | 0.01 |
ENSRNOT00000092092
|
AABR07017236.1
|
|
| chr13_+_92264231 | 0.01 |
ENSRNOT00000066509
ENSRNOT00000004716 |
Spta1
|
spectrin, alpha, erythrocytic 1 |
| chr3_+_101899068 | 0.01 |
ENSRNOT00000079600
ENSRNOT00000006256 |
Muc15
|
mucin 15, cell surface associated |
| chr11_-_69223158 | 0.01 |
ENSRNOT00000067060
|
Mylk
|
myosin light chain kinase |
| chr1_-_23556241 | 0.01 |
ENSRNOT00000072943
|
LOC100910446
|
syntaxin-7-like |
| chrX_-_63291107 | 0.01 |
ENSRNOT00000092019
|
Eif2s3
|
eukaryotic translation initiation factor 2 subunit gamma |
| chr6_-_26546545 | 0.01 |
ENSRNOT00000091264
|
Snx17
|
sorting nexin 17 |
| chr10_-_43760930 | 0.01 |
ENSRNOT00000003639
|
Zfp672
|
zinc finger protein 672 |
| chr7_+_13318533 | 0.01 |
ENSRNOT00000034545
|
LOC691170
|
similar to zinc finger protein 84 (HPF2) |
| chr2_-_205212681 | 0.01 |
ENSRNOT00000022575
|
Tshb
|
thyroid stimulating hormone, beta |
| chr2_-_227145185 | 0.01 |
ENSRNOT00000083189
|
AABR07013202.1
|
|
| chr20_-_7142213 | 0.01 |
ENSRNOT00000078918
|
RGD1564450
|
RGD1564450 |
| chr15_+_93612971 | 0.00 |
ENSRNOT00000013182
|
Acod1
|
aconitate decarboxylase 1 |
| chr10_+_109786222 | 0.00 |
ENSRNOT00000054953
|
Npb
|
neuropeptide B |
| chr13_+_44957014 | 0.00 |
ENSRNOT00000004878
|
Ubxn4
|
UBX domain protein 4 |
| chr1_-_80271001 | 0.00 |
ENSRNOT00000034266
|
Cd3eap
|
CD3e molecule associated protein |
| chr2_-_88553086 | 0.00 |
ENSRNOT00000042494
|
LOC361914
|
similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 |
| chr15_+_32386816 | 0.00 |
ENSRNOT00000060322
|
AABR07017901.1
|
|
| chr3_-_83048289 | 0.00 |
ENSRNOT00000047571
ENSRNOT00000012806 |
Hsd17b12
|
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr13_+_73708815 | 0.00 |
ENSRNOT00000071482
ENSRNOT00000077874 ENSRNOT00000050734 |
Tor1aip2
Tor1aip2
|
torsin 1A interacting protein 2 torsin 1A interacting protein 2 |
| chr2_-_34254968 | 0.00 |
ENSRNOT00000016439
|
Trappc13
|
trafficking protein particle complex 13 |
| chr5_+_98387291 | 0.00 |
ENSRNOT00000046503
|
Tyrp1
|
tyrosinase-related protein 1 |
| chr10_-_25890639 | 0.00 |
ENSRNOT00000085499
|
Hmmr
|
hyaluronan-mediated motility receptor |
| chr20_-_29558321 | 0.00 |
ENSRNOT00000000702
|
Anapc16
|
anaphase promoting complex subunit 16 |
| chr17_+_30670171 | 0.00 |
ENSRNOT00000022369
|
Fam217a
|
family with sequence similarity 217, member A |
| chr10_-_25847994 | 0.00 |
ENSRNOT00000082076
|
Mat2b
|
methionine adenosyltransferase 2B |
| chr10_-_65424802 | 0.00 |
ENSRNOT00000018468
|
Traf4
|
Tnf receptor associated factor 4 |
| chr20_-_45260119 | 0.00 |
ENSRNOT00000000718
|
Slc16a10
|
solute carrier family 16 member 10 |
| chr6_+_97110158 | 0.00 |
ENSRNOT00000088430
|
Syt16
|
synaptotagmin 16 |
| chr10_+_53595854 | 0.00 |
ENSRNOT00000044116
|
Sco1
|
SCO1 cytochrome c oxidase assembly protein |
| chr3_+_159995064 | 0.00 |
ENSRNOT00000012606
|
Ttpal
|
alpha tocopherol transfer protein like |
| chr10_-_53595827 | 0.00 |
ENSRNOT00000004560
|
Adprm
|
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax6
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0021917 | pancreatic A cell development(GO:0003322) forebrain-midbrain boundary formation(GO:0021905) somatic motor neuron fate commitment(GO:0021917) regulation of transcription from RNA polymerase II promoter involved in somatic motor neuron fate commitment(GO:0021918) sensory neuron migration(GO:1904937) |
| 0.0 | 0.1 | GO:0006045 | N-acetylglucosamine biosynthetic process(GO:0006045) N-acetylneuraminate biosynthetic process(GO:0046380) |
| 0.0 | 0.0 | GO:0045575 | basophil activation(GO:0045575) |
| 0.0 | 0.0 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:0001574 | ganglioside biosynthetic process(GO:0001574) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.0 | GO:0035838 | growing cell tip(GO:0035838) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003828 | alpha-N-acetylneuraminate alpha-2,8-sialyltransferase activity(GO:0003828) |
| 0.0 | 0.1 | GO:0008311 | double-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008311) |
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.1 | GO:0009384 | N-acylmannosamine kinase activity(GO:0009384) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.0 | 0.0 | GO:0015057 | thrombin receptor activity(GO:0015057) |