Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
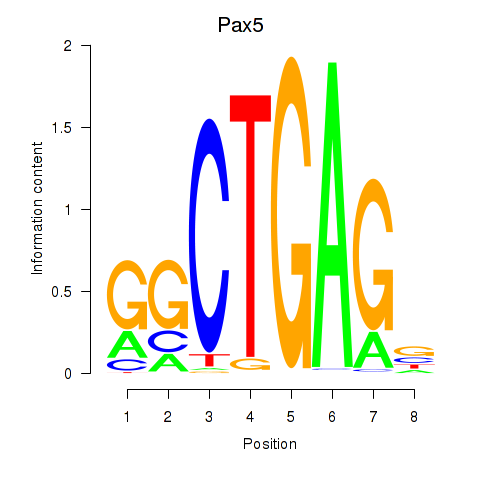
Results for Pax5
Z-value: 1.11
Transcription factors associated with Pax5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax5
|
ENSRNOG00000024729 | paired box 5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax5 | rn6_v1_chr5_-_60191941_60191941 | 0.23 | 7.1e-01 | Click! |
Activity profile of Pax5 motif
Sorted Z-values of Pax5 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_150167077 | 0.85 |
ENSRNOT00000088493
ENSRNOT00000013761 ENSRNOT00000067349 ENSRNOT00000051097 ENSRNOT00000067189 |
Epb41
|
erythrocyte membrane protein band 4.1 |
| chr8_-_98738446 | 0.80 |
ENSRNOT00000019860
|
Zic1
|
Zic family member 1 |
| chr7_-_3342491 | 0.78 |
ENSRNOT00000081756
|
Rdh5
|
retinol dehydrogenase 5 |
| chr20_+_5008508 | 0.73 |
ENSRNOT00000001153
|
Vwa7
|
von Willebrand factor A domain containing 7 |
| chr2_-_259150479 | 0.71 |
ENSRNOT00000085892
|
AABR07013843.1
|
|
| chr6_-_27190126 | 0.62 |
ENSRNOT00000068412
ENSRNOT00000013107 |
Kcnk3
|
potassium two pore domain channel subfamily K member 3 |
| chr6_-_11494459 | 0.54 |
ENSRNOT00000021570
|
Kcnk12
|
potassium two pore domain channel subfamily K member 12 |
| chr10_-_88551056 | 0.49 |
ENSRNOT00000032737
|
Zfp385c
|
zinc finger protein 385C |
| chr10_-_55642681 | 0.48 |
ENSRNOT00000057157
|
AC129753.1
|
|
| chr8_+_118066988 | 0.48 |
ENSRNOT00000056161
|
Map4
|
microtubule-associated protein 4 |
| chr8_-_115167486 | 0.45 |
ENSRNOT00000033018
|
Gpr62
|
G protein-coupled receptor 62 |
| chr6_+_110624856 | 0.45 |
ENSRNOT00000014017
|
Vash1
|
vasohibin 1 |
| chr5_-_156811650 | 0.44 |
ENSRNOT00000068065
|
Fam43b
|
family with sequence similarity 43, member B |
| chr19_+_55669626 | 0.43 |
ENSRNOT00000033352
|
Cdh15
|
cadherin 15 |
| chr17_-_23923792 | 0.42 |
ENSRNOT00000018739
|
Gfod1
|
glucose-fructose oxidoreductase domain containing 1 |
| chr20_+_4823662 | 0.41 |
ENSRNOT00000090028
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr18_+_27558089 | 0.40 |
ENSRNOT00000027499
|
Fam53c
|
family with sequence similarity 53, member C |
| chr13_-_49313940 | 0.39 |
ENSRNOT00000012190
|
Cntn2
|
contactin 2 |
| chr4_+_148139528 | 0.39 |
ENSRNOT00000092594
ENSRNOT00000015620 ENSRNOT00000092613 |
Washc2c
|
WASH complex subunit 2C |
| chr11_+_57207656 | 0.36 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr20_-_4823475 | 0.36 |
ENSRNOT00000082536
ENSRNOT00000001114 |
Atp6v1g2
|
ATPase H+ transporting V1 subunit G2 |
| chr20_+_3558827 | 0.36 |
ENSRNOT00000088130
|
Ddr1
|
discoidin domain receptor tyrosine kinase 1 |
| chr13_+_71107465 | 0.35 |
ENSRNOT00000003239
|
Rgs8
|
regulator of G-protein signaling 8 |
| chr2_+_195617021 | 0.34 |
ENSRNOT00000067042
ENSRNOT00000036656 |
Rorc
Lingo4
|
RAR-related orphan receptor C leucine rich repeat and Ig domain containing 4 |
| chr12_+_28212333 | 0.33 |
ENSRNOT00000001182
|
Auts2
|
autism susceptibility candidate 2 |
| chrX_+_105239840 | 0.33 |
ENSRNOT00000039864
|
Drp2
|
dystrophin related protein 2 |
| chr3_+_80676820 | 0.32 |
ENSRNOT00000084809
|
Ambra1
|
autophagy and beclin 1 regulator 1 |
| chr11_+_71151132 | 0.31 |
ENSRNOT00000082594
ENSRNOT00000082435 |
Rubcn
|
RUN and cysteine rich domain containing beclin 1 interacting protein |
| chr1_+_264504591 | 0.31 |
ENSRNOT00000050076
|
Pax2
|
paired box 2 |
| chr17_-_10818835 | 0.31 |
ENSRNOT00000091046
|
Cplx2
|
complexin 2 |
| chr1_+_82253005 | 0.30 |
ENSRNOT00000027854
|
AC121639.1
|
|
| chr20_-_3386048 | 0.30 |
ENSRNOT00000089550
ENSRNOT00000065025 |
Dhx16
|
DEAH-box helicase 16 |
| chr14_+_83724933 | 0.30 |
ENSRNOT00000029848
|
Pla2g3
|
phospholipase A2, group III |
| chr7_-_119441487 | 0.30 |
ENSRNOT00000067635
|
Pvalb
|
parvalbumin |
| chr5_+_169475270 | 0.29 |
ENSRNOT00000014625
|
Icmt
|
isoprenylcysteine carboxyl methyltransferase |
| chr1_+_33910912 | 0.29 |
ENSRNOT00000044690
|
Irx1
|
iroquois homeobox 1 |
| chr6_-_111222858 | 0.29 |
ENSRNOT00000074707
|
Tmed8
|
transmembrane p24 trafficking protein 8 |
| chr3_+_147609095 | 0.28 |
ENSRNOT00000041456
|
Srxn1
|
sulfiredoxin 1 |
| chr10_-_62184874 | 0.28 |
ENSRNOT00000004229
|
Rpa1
|
replication protein A1 |
| chr3_+_162692185 | 0.28 |
ENSRNOT00000007768
|
Ncoa3
|
nuclear receptor coactivator 3 |
| chr9_+_117795132 | 0.28 |
ENSRNOT00000086943
|
Akain1
|
A-kinase anchor inhibitor 1 |
| chr10_+_78168715 | 0.28 |
ENSRNOT00000072294
|
Cox11
|
COX11 cytochrome c oxidase copper chaperone |
| chr5_-_166133491 | 0.27 |
ENSRNOT00000087739
ENSRNOT00000089099 |
Kif1b
|
kinesin family member 1B |
| chr15_-_33775109 | 0.27 |
ENSRNOT00000033722
|
Jph4
|
junctophilin 4 |
| chr1_-_215846911 | 0.27 |
ENSRNOT00000089171
|
Igf2
|
insulin-like growth factor 2 |
| chr1_-_5165859 | 0.27 |
ENSRNOT00000044325
ENSRNOT00000019319 |
Grm1
|
glutamate metabotropic receptor 1 |
| chr3_-_160301552 | 0.26 |
ENSRNOT00000014498
|
Rims4
|
regulating synaptic membrane exocytosis 4 |
| chr10_+_109278712 | 0.26 |
ENSRNOT00000065565
|
LOC690871
|
hypothetical protein LOC690871 |
| chr1_+_199449973 | 0.26 |
ENSRNOT00000029994
|
Trim72
|
tripartite motif containing 72 |
| chr3_+_11198401 | 0.26 |
ENSRNOT00000088576
ENSRNOT00000066368 |
Prrc2b
|
proline-rich coiled-coil 2B |
| chr6_-_122897997 | 0.26 |
ENSRNOT00000057601
|
Eml5
|
echinoderm microtubule associated protein like 5 |
| chr4_+_168599331 | 0.26 |
ENSRNOT00000086719
|
Crebl2
|
cAMP responsive element binding protein-like 2 |
| chr5_+_148661070 | 0.25 |
ENSRNOT00000056229
|
Nkain1
|
Sodium/potassium transporting ATPase interacting 1 |
| chr6_+_24163026 | 0.25 |
ENSRNOT00000061284
|
Lbh
|
limb bud and heart development |
| chr16_-_48437223 | 0.25 |
ENSRNOT00000013005
ENSRNOT00000059401 |
Enpp6
|
ectonucleotide pyrophosphatase/phosphodiesterase 6 |
| chr10_-_64657089 | 0.25 |
ENSRNOT00000080703
|
Abr
|
active BCR-related |
| chr15_-_44860604 | 0.25 |
ENSRNOT00000018637
|
Nefm
|
neurofilament medium |
| chr20_+_4822935 | 0.24 |
ENSRNOT00000077200
|
Nfkbil1
|
NFKB inhibitor like 1 |
| chr10_-_74679858 | 0.24 |
ENSRNOT00000003859
|
Ppm1e
|
protein phosphatase, Mg2+/Mn2+ dependent, 1E |
| chr20_+_29655226 | 0.24 |
ENSRNOT00000089059
|
Spock2
|
SPARC/osteonectin, cwcv and kazal like domains proteoglycan 2 |
| chr12_-_11783232 | 0.24 |
ENSRNOT00000075888
|
Zfp498
|
zinc finger protein 498 |
| chr7_-_117364697 | 0.24 |
ENSRNOT00000077314
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr10_-_62483342 | 0.24 |
ENSRNOT00000079956
|
LOC100912068
|
hypermethylated in cancer 1 protein-like |
| chr1_-_57815038 | 0.23 |
ENSRNOT00000075401
|
Rgmb
|
repulsive guidance molecule family member B |
| chr17_-_27112820 | 0.23 |
ENSRNOT00000018359
|
Bmp6
|
bone morphogenetic protein 6 |
| chr10_+_63829807 | 0.23 |
ENSRNOT00000006407
|
Crk
|
CRK proto-oncogene, adaptor protein |
| chr1_-_209641123 | 0.23 |
ENSRNOT00000021702
|
Ebf3
|
early B-cell factor 3 |
| chr16_-_10726648 | 0.23 |
ENSRNOT00000080635
ENSRNOT00000087150 |
Sncg
|
synuclein, gamma |
| chr1_+_165625350 | 0.23 |
ENSRNOT00000087534
|
Rab6a
|
RAB6A, member RAS oncogene family |
| chr7_-_117364322 | 0.22 |
ENSRNOT00000080724
|
Oplah
|
5-oxoprolinase (ATP-hydrolysing) |
| chr10_+_56381813 | 0.22 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr16_-_81945127 | 0.22 |
ENSRNOT00000023352
|
Mcf2l
|
MCF.2 cell line derived transforming sequence-like |
| chr2_-_32518643 | 0.22 |
ENSRNOT00000061032
|
Mast4
|
microtubule associated serine/threonine kinase family member 4 |
| chr7_-_11824742 | 0.22 |
ENSRNOT00000051467
|
Dot1l
|
DOT1 like histone lysine methyltransferase |
| chr14_+_83343330 | 0.21 |
ENSRNOT00000089198
|
Pisd
|
phosphatidylserine decarboxylase |
| chr19_-_19832812 | 0.21 |
ENSRNOT00000084657
|
Papd5
|
poly(A) RNA polymerase D5, non-canonical |
| chr1_-_100669684 | 0.21 |
ENSRNOT00000091760
|
Myh14
|
myosin heavy chain 14 |
| chr4_-_150471806 | 0.21 |
ENSRNOT00000008741
ENSRNOT00000076557 |
Bms1
|
BMS1 ribosome biogenesis factor |
| chr13_-_51183269 | 0.21 |
ENSRNOT00000039540
|
Ppfia4
|
PTPRF interacting protein alpha 4 |
| chr3_+_93495106 | 0.21 |
ENSRNOT00000029922
ENSRNOT00000085760 |
Abtb2
|
ankyrin repeat and BTB domain containing 2 |
| chr1_+_82174451 | 0.20 |
ENSRNOT00000027783
|
Tmem145
|
transmembrane protein 145 |
| chr1_+_154606490 | 0.20 |
ENSRNOT00000024095
|
Ccdc89
|
coiled-coil domain containing 89 |
| chr3_-_110492398 | 0.20 |
ENSRNOT00000056450
|
Ankrd63
|
ankyrin repeat domain 63 |
| chrX_+_105500173 | 0.20 |
ENSRNOT00000040476
|
Armcx4
|
armadillo repeat containing, X-linked 4 |
| chr15_-_108898703 | 0.20 |
ENSRNOT00000067577
|
Zic5
|
Zic family member 5 |
| chr5_+_76092287 | 0.20 |
ENSRNOT00000020207
|
Zfp483
|
zinc finger protein 483 |
| chr20_+_5535432 | 0.20 |
ENSRNOT00000040859
|
Syngap1
|
synaptic Ras GTPase activating protein 1 |
| chr1_-_198316882 | 0.20 |
ENSRNOT00000085304
ENSRNOT00000064985 |
Taok2
|
TAO kinase 2 |
| chr20_-_31597830 | 0.20 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr6_-_126582034 | 0.19 |
ENSRNOT00000010656
ENSRNOT00000080829 |
Itpk1
|
inositol-tetrakisphosphate 1-kinase |
| chr10_+_56662561 | 0.19 |
ENSRNOT00000025254
|
Asgr1
|
asialoglycoprotein receptor 1 |
| chr1_-_281874456 | 0.19 |
ENSRNOT00000084760
ENSRNOT00000050617 |
Cacul1
|
CDK2-associated, cullin domain 1 |
| chr10_+_55707164 | 0.19 |
ENSRNOT00000009757
|
Hes7
|
hes family bHLH transcription factor 7 |
| chr18_+_55797198 | 0.19 |
ENSRNOT00000026334
ENSRNOT00000026394 |
Dctn4
|
dynactin subunit 4 |
| chr1_+_221612584 | 0.18 |
ENSRNOT00000090100
|
Atg2a
|
autophagy related 2A |
| chr7_-_117978787 | 0.18 |
ENSRNOT00000074262
|
Zfp647
|
zinc finger protein 647 |
| chr7_+_70364813 | 0.18 |
ENSRNOT00000084012
ENSRNOT00000031230 |
Agap2
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chr1_+_199196059 | 0.18 |
ENSRNOT00000090428
|
Fbxl19
|
F-box and leucine-rich repeat protein 19 |
| chr7_-_143793774 | 0.18 |
ENSRNOT00000079678
|
Csad
|
cysteine sulfinic acid decarboxylase |
| chr13_+_105684420 | 0.18 |
ENSRNOT00000040543
|
Gpatch2
|
G patch domain containing 2 |
| chr2_-_174413236 | 0.18 |
ENSRNOT00000064588
|
Golim4
|
golgi integral membrane protein 4 |
| chr2_+_207930796 | 0.18 |
ENSRNOT00000047827
|
Kcnd3
|
potassium voltage-gated channel subfamily D member 3 |
| chr7_-_24313885 | 0.18 |
ENSRNOT00000044331
|
Btbd11
|
BTB domain containing 11 |
| chr20_+_7279820 | 0.18 |
ENSRNOT00000090820
|
Pacsin1
|
protein kinase C and casein kinase substrate in neurons 1 |
| chr1_-_43638161 | 0.17 |
ENSRNOT00000024460
|
Ipcef1
|
interaction protein for cytohesin exchange factors 1 |
| chr11_-_83926524 | 0.17 |
ENSRNOT00000041777
ENSRNOT00000040029 |
Eif4g1
|
eukaryotic translation initiation factor 4 gamma, 1 |
| chr15_+_31243097 | 0.17 |
ENSRNOT00000042721
|
LOC100360891
|
RGD1359684 protein-like |
| chr2_+_41157823 | 0.17 |
ENSRNOT00000066384
|
Pde4d
|
phosphodiesterase 4D |
| chr16_-_8685529 | 0.17 |
ENSRNOT00000092751
|
Slc18a3
|
solute carrier family 18 member A3 |
| chr5_+_142875773 | 0.17 |
ENSRNOT00000082120
ENSRNOT00000056496 |
Epha10
|
EPH receptor A10 |
| chr14_-_84170301 | 0.16 |
ENSRNOT00000080413
|
Slc35e4
|
solute carrier family 35, member E4 |
| chr4_-_115015965 | 0.16 |
ENSRNOT00000014603
|
Mthfd2
|
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr1_+_226252226 | 0.16 |
ENSRNOT00000039369
|
AABR07006258.1
|
|
| chr7_-_3074359 | 0.16 |
ENSRNOT00000019738
|
Ikzf4
|
IKAROS family zinc finger 4 |
| chr14_+_84876781 | 0.16 |
ENSRNOT00000065188
|
Ascc2
|
activating signal cointegrator 1 complex subunit 2 |
| chr8_+_109455786 | 0.16 |
ENSRNOT00000039593
|
Msl2
|
male-specific lethal 2 homolog (Drosophila) |
| chr4_+_26470864 | 0.15 |
ENSRNOT00000021979
|
Fzd1
|
frizzled class receptor 1 |
| chr9_+_119542328 | 0.15 |
ENSRNOT00000082005
|
Lpin2
|
lipin 2 |
| chr19_-_1074333 | 0.15 |
ENSRNOT00000017983
ENSRNOT00000086995 |
Cdh5
|
cadherin 5 |
| chr6_+_24160226 | 0.15 |
ENSRNOT00000091904
|
Lbh
|
limb bud and heart development |
| chr5_+_58855773 | 0.15 |
ENSRNOT00000072869
|
Rusc2
|
RUN and SH3 domain containing 2 |
| chr3_+_61658245 | 0.15 |
ENSRNOT00000033511
|
Hoxd3
|
homeo box D3 |
| chr1_-_192088520 | 0.15 |
ENSRNOT00000047420
|
Palb2
|
partner and localizer of BRCA2 |
| chr14_-_55081551 | 0.15 |
ENSRNOT00000049245
|
Pcdh7
|
protocadherin 7 |
| chr9_+_94425252 | 0.15 |
ENSRNOT00000064965
ENSRNOT00000076099 |
Gigyf2
|
GRB10 interacting GYF protein 2 |
| chr7_-_98709344 | 0.14 |
ENSRNOT00000064122
|
Tmem65
|
transmembrane protein 65 |
| chr2_-_211831551 | 0.14 |
ENSRNOT00000039225
ENSRNOT00000090561 |
Fam102b
|
family with sequence similarity 102, member B |
| chr10_+_36098051 | 0.14 |
ENSRNOT00000083971
|
Adamts2
|
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr12_-_5773036 | 0.14 |
ENSRNOT00000041365
|
Fry
|
FRY microtubule binding protein |
| chr17_-_18421861 | 0.14 |
ENSRNOT00000060500
ENSRNOT00000036560 |
Fam8a1
|
family with sequence similarity 8, member A1 |
| chr7_-_67116980 | 0.14 |
ENSRNOT00000005798
|
Ppm1h
|
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chrX_-_2657155 | 0.14 |
ENSRNOT00000005630
|
Chst7
|
carbohydrate sulfotransferase 7 |
| chr1_-_226791773 | 0.14 |
ENSRNOT00000082482
ENSRNOT00000065376 ENSRNOT00000054812 ENSRNOT00000086669 |
LOC100911215
|
T-cell surface glycoprotein CD5-like |
| chr1_+_198932870 | 0.14 |
ENSRNOT00000055003
|
Fbrs
|
fibrosin |
| chr12_+_2589818 | 0.14 |
ENSRNOT00000087848
|
AABR07034980.1
|
|
| chr4_-_157274755 | 0.14 |
ENSRNOT00000088895
|
LOC100911672
|
atrophin-1-like |
| chr10_-_56444847 | 0.14 |
ENSRNOT00000056872
ENSRNOT00000092662 |
Nlgn2
|
neuroligin 2 |
| chr2_+_187737770 | 0.14 |
ENSRNOT00000036143
ENSRNOT00000092611 ENSRNOT00000092620 |
Paqr6
|
progestin and adipoQ receptor family member 6 |
| chr1_-_198460126 | 0.14 |
ENSRNOT00000082940
ENSRNOT00000086019 |
Maz
|
MYC associated zinc finger protein |
| chr2_-_174413038 | 0.14 |
ENSRNOT00000089229
ENSRNOT00000037115 |
Golim4
|
golgi integral membrane protein 4 |
| chr5_-_65073012 | 0.14 |
ENSRNOT00000007957
|
Grin3a
|
glutamate ionotropic receptor NMDA type subunit 3A |
| chr14_+_42714315 | 0.14 |
ENSRNOT00000084095
ENSRNOT00000091449 |
Phox2b
|
paired-like homeobox 2b |
| chr1_-_174119815 | 0.13 |
ENSRNOT00000019368
|
Trim66
|
tripartite motif-containing 66 |
| chr17_+_5225835 | 0.13 |
ENSRNOT00000022373
|
Zcchc6
|
zinc finger CCHC-type containing 6 |
| chr17_+_77185053 | 0.13 |
ENSRNOT00000091561
|
Optn
|
optineurin |
| chr19_+_11473541 | 0.13 |
ENSRNOT00000082066
|
Amfr
|
autocrine motility factor receptor |
| chr16_+_7588217 | 0.13 |
ENSRNOT00000026525
|
Eaf1
|
ELL associated factor 1 |
| chr5_-_168734296 | 0.13 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr1_-_93949187 | 0.13 |
ENSRNOT00000018956
|
Zfp536
|
zinc finger protein 536 |
| chr5_+_144779681 | 0.13 |
ENSRNOT00000016493
|
LOC100294508
|
dyslexia susceptibility 2-like |
| chr4_+_168752133 | 0.13 |
ENSRNOT00000010289
|
Apold1
|
apolipoprotein L domain containing 1 |
| chr6_+_99625306 | 0.13 |
ENSRNOT00000008573
|
Plekhg3
|
pleckstrin homology and RhoGEF domain containing G3 |
| chr17_-_10208360 | 0.13 |
ENSRNOT00000087397
|
Unc5a
|
unc-5 netrin receptor A |
| chr3_-_80003032 | 0.13 |
ENSRNOT00000017360
|
Madd
|
MAP-kinase activating death domain |
| chr10_-_55923964 | 0.13 |
ENSRNOT00000073296
|
Cntrob
|
centrobin, centriole duplication and spindle assembly protein |
| chr1_+_167374182 | 0.13 |
ENSRNOT00000092882
|
Stim1
|
stromal interaction molecule 1 |
| chr6_+_108285822 | 0.13 |
ENSRNOT00000015889
|
Vsx2
|
visual system homeobox 2 |
| chr15_+_52241801 | 0.12 |
ENSRNOT00000082639
|
Hr
|
HR, lysine demethylase and nuclear receptor corepressor |
| chr19_-_38484611 | 0.12 |
ENSRNOT00000077960
|
Nfat5
|
nuclear factor of activated T-cells 5 |
| chr13_-_51784639 | 0.12 |
ENSRNOT00000089068
|
Ppp1r12b
|
protein phosphatase 1, regulatory subunit 12B |
| chr8_+_29453643 | 0.12 |
ENSRNOT00000090643
|
Opcml
|
opioid binding protein/cell adhesion molecule-like |
| chr1_-_14412807 | 0.12 |
ENSRNOT00000074583
|
Tnfaip3
|
TNF alpha induced protein 3 |
| chr2_-_62634785 | 0.12 |
ENSRNOT00000017937
|
Pdzd2
|
PDZ domain containing 2 |
| chr20_-_13706205 | 0.12 |
ENSRNOT00000038623
|
Derl3
|
derlin 3 |
| chr2_-_63166509 | 0.12 |
ENSRNOT00000018246
|
Cdh6
|
cadherin 6 |
| chrX_+_71272042 | 0.12 |
ENSRNOT00000076034
ENSRNOT00000076816 |
Gjb1
|
gap junction protein, beta 1 |
| chr13_-_89267402 | 0.12 |
ENSRNOT00000004179
|
Dusp12
|
dual specificity phosphatase 12 |
| chr16_+_23668595 | 0.12 |
ENSRNOT00000067886
|
Psd3
|
pleckstrin and Sec7 domain containing 3 |
| chr8_-_61079526 | 0.12 |
ENSRNOT00000068658
|
Peak1
|
pseudopodium-enriched atypical kinase 1 |
| chr2_+_34186091 | 0.11 |
ENSRNOT00000016129
|
Sgtb
|
small glutamine rich tetratricopeptide repeat containing beta |
| chr3_-_148312420 | 0.11 |
ENSRNOT00000047416
ENSRNOT00000081272 |
Bcl2l1
|
Bcl2-like 1 |
| chr2_-_206274079 | 0.11 |
ENSRNOT00000056079
|
Hipk1
|
homeodomain interacting protein kinase 1 |
| chr10_+_70689863 | 0.11 |
ENSRNOT00000091122
ENSRNOT00000086987 ENSRNOT00000082030 |
Taf15
|
TATA-box binding protein associated factor 15 |
| chr11_-_73678984 | 0.11 |
ENSRNOT00000002355
|
Fam43a
|
family with sequence similarity 43, member A |
| chrX_-_1714061 | 0.11 |
ENSRNOT00000011592
|
Cdk16
|
cyclin-dependent kinase 16 |
| chr4_+_64088900 | 0.11 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr14_+_115275894 | 0.11 |
ENSRNOT00000033437
|
Gpr75
|
G protein-coupled receptor 75 |
| chr16_+_10727571 | 0.11 |
ENSRNOT00000084422
|
Mmrn2
|
multimerin 2 |
| chr1_-_219745654 | 0.11 |
ENSRNOT00000054848
|
LOC689065
|
hypothetical protein LOC689065 |
| chr5_+_169181418 | 0.11 |
ENSRNOT00000004508
|
Klhl21
|
kelch-like family member 21 |
| chr18_+_30574627 | 0.11 |
ENSRNOT00000060484
|
Pcdhb19
|
protocadherin beta 19 |
| chr13_-_52266100 | 0.11 |
ENSRNOT00000087532
|
AC096239.3
|
|
| chrX_+_23081125 | 0.11 |
ENSRNOT00000071639
|
AABR07037520.1
|
|
| chr3_+_12262822 | 0.11 |
ENSRNOT00000022585
|
Angptl2
|
angiopoietin-like 2 |
| chr1_-_264756546 | 0.11 |
ENSRNOT00000020020
|
Mrpl43
|
mitochondrial ribosomal protein L43 |
| chr15_+_30429974 | 0.11 |
ENSRNOT00000071797
|
AABR07017735.1
|
|
| chr3_+_55094637 | 0.11 |
ENSRNOT00000058763
|
Cers6
|
ceramide synthase 6 |
| chr20_+_11972381 | 0.10 |
ENSRNOT00000001642
|
Adarb1
|
adenosine deaminase, RNA-specific, B1 |
| chr7_-_122403667 | 0.10 |
ENSRNOT00000088814
|
Mkl1
|
megakaryoblastic leukemia (translocation) 1 |
| chr6_+_137997335 | 0.10 |
ENSRNOT00000006872
|
Tmem121
|
transmembrane protein 121 |
| chr7_-_116255167 | 0.10 |
ENSRNOT00000038109
ENSRNOT00000041774 |
Cyp11b2
|
cytochrome P450, family 11, subfamily b, polypeptide 2 |
| chr7_+_2752680 | 0.10 |
ENSRNOT00000033726
|
Cs
|
citrate synthase |
| chr9_+_16647922 | 0.10 |
ENSRNOT00000031625
|
Klc4
|
kinesin light chain 4 |
| chr14_-_84393421 | 0.10 |
ENSRNOT00000006911
|
Ccdc157
|
coiled-coil domain containing 157 |
| chr10_+_45258887 | 0.10 |
ENSRNOT00000048642
|
Btnl10
|
butyrophilin like 10 |
| chr2_-_186515135 | 0.10 |
ENSRNOT00000077375
|
Kirrel
|
kin of IRRE like (Drosophila) |
| chrX_-_15707436 | 0.10 |
ENSRNOT00000085907
|
Syp
|
synaptophysin |
| chr1_-_127599257 | 0.10 |
ENSRNOT00000018436
|
Asb7
|
ankyrin repeat and SOCS box-containing 7 |
| chr10_+_108750620 | 0.10 |
ENSRNOT00000005337
|
Rptor
|
regulatory associated protein of MTOR, complex 1 |
| chr10_-_74769637 | 0.10 |
ENSRNOT00000008889
|
LOC102555183
|
zinc finger protein OZF-like |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax5
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.4 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.1 | 0.4 | GO:1901491 | negative regulation of lymphangiogenesis(GO:1901491) |
| 0.1 | 0.3 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.3 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.1 | 0.3 | GO:0035566 | regulation of metanephros size(GO:0035566) |
| 0.1 | 0.4 | GO:2000813 | negative regulation of barbed-end actin filament capping(GO:2000813) |
| 0.1 | 0.8 | GO:0031665 | negative regulation of lipopolysaccharide-mediated signaling pathway(GO:0031665) |
| 0.1 | 0.3 | GO:0043314 | negative regulation of neutrophil degranulation(GO:0043314) |
| 0.1 | 0.2 | GO:0033693 | neurofilament bundle assembly(GO:0033693) |
| 0.1 | 0.3 | GO:0072268 | pattern specification involved in metanephros development(GO:0072268) |
| 0.1 | 0.3 | GO:0033132 | negative regulation of glucokinase activity(GO:0033132) negative regulation of hexokinase activity(GO:1903300) |
| 0.1 | 0.4 | GO:1904674 | positive regulation of somatic stem cell population maintenance(GO:1904674) |
| 0.1 | 0.2 | GO:0042412 | taurine biosynthetic process(GO:0042412) |
| 0.1 | 0.2 | GO:0032346 | positive regulation of aldosterone metabolic process(GO:0032346) positive regulation of aldosterone biosynthetic process(GO:0032349) positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.1 | 0.2 | GO:1990859 | response to endothelin(GO:1990839) cellular response to endothelin(GO:1990859) |
| 0.1 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.3 | GO:0072615 | interleukin-17 secretion(GO:0072615) |
| 0.1 | 0.2 | GO:0034729 | histone H3-K79 methylation(GO:0034729) |
| 0.1 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.3 | GO:0019521 | aldonic acid metabolic process(GO:0019520) D-gluconate metabolic process(GO:0019521) |
| 0.0 | 0.6 | GO:0071294 | cellular response to zinc ion(GO:0071294) |
| 0.0 | 0.3 | GO:0042695 | thelarche(GO:0042695) dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) mammary gland branching involved in thelarche(GO:0060744) |
| 0.0 | 0.1 | GO:0001966 | thigmotaxis(GO:0001966) |
| 0.0 | 0.2 | GO:0071464 | cellular response to hydrostatic pressure(GO:0071464) |
| 0.0 | 0.1 | GO:0048880 | medulla oblongata development(GO:0021550) sensory system development(GO:0048880) |
| 0.0 | 0.3 | GO:0046813 | receptor-mediated virion attachment to host cell(GO:0046813) |
| 0.0 | 0.1 | GO:0036395 | pancreatic amylase secretion(GO:0036395) regulation of pancreatic amylase secretion(GO:1902276) |
| 0.0 | 0.3 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.1 | GO:1905218 | cellular response to astaxanthin(GO:1905218) |
| 0.0 | 0.1 | GO:0021615 | glossopharyngeal nerve morphogenesis(GO:0021615) |
| 0.0 | 0.1 | GO:1904428 | negative regulation of tubulin deacetylation(GO:1904428) |
| 0.0 | 0.1 | GO:0061744 | motor behavior(GO:0061744) |
| 0.0 | 0.1 | GO:0006369 | termination of RNA polymerase II transcription(GO:0006369) |
| 0.0 | 0.1 | GO:2000474 | regulation of opioid receptor signaling pathway(GO:2000474) |
| 0.0 | 0.1 | GO:1903233 | regulation of calcium ion-dependent exocytosis of neurotransmitter(GO:1903233) calcium ion import into sarcoplasmic reticulum(GO:1990036) |
| 0.0 | 0.1 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.3 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.0 | 0.2 | GO:0019800 | peptide cross-linking via chondroitin 4-sulfate glycosaminoglycan(GO:0019800) |
| 0.0 | 0.1 | GO:0000448 | cleavage in ITS2 between 5.8S rRNA and LSU-rRNA of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000448) |
| 0.0 | 0.1 | GO:0071205 | protein localization to juxtaparanode region of axon(GO:0071205) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.4 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0071579 | regulation of zinc ion transport(GO:0071579) |
| 0.0 | 0.4 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.1 | GO:0001982 | baroreceptor response to decreased systemic arterial blood pressure(GO:0001982) |
| 0.0 | 0.2 | GO:1903142 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.2 | GO:2000507 | positive regulation of energy homeostasis(GO:2000507) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.3 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.3 | GO:0031915 | positive regulation of synaptic plasticity(GO:0031915) |
| 0.0 | 0.1 | GO:0016344 | meiotic chromosome movement towards spindle pole(GO:0016344) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.2 | GO:1902410 | mitotic cytokinetic process(GO:1902410) |
| 0.0 | 0.3 | GO:0046498 | S-adenosylhomocysteine metabolic process(GO:0046498) |
| 0.0 | 0.4 | GO:0071044 | histone mRNA catabolic process(GO:0071044) |
| 0.0 | 0.3 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0002175 | protein localization to paranode region of axon(GO:0002175) |
| 0.0 | 0.2 | GO:0019695 | choline metabolic process(GO:0019695) |
| 0.0 | 0.3 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.2 | GO:0014807 | regulation of somitogenesis(GO:0014807) |
| 0.0 | 0.1 | GO:0048387 | negative regulation of retinoic acid receptor signaling pathway(GO:0048387) |
| 0.0 | 0.1 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) negative regulation of adipose tissue development(GO:1904178) |
| 0.0 | 0.7 | GO:0030866 | cortical actin cytoskeleton organization(GO:0030866) |
| 0.0 | 0.0 | GO:0080033 | cellular response to nitrite(GO:0071250) response to nitrite(GO:0080033) |
| 0.0 | 0.1 | GO:1903544 | cellular response to interleukin-11(GO:0071348) response to butyrate(GO:1903544) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.1 | GO:0002432 | granuloma formation(GO:0002432) |
| 0.0 | 0.1 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0033131 | regulation of glucokinase activity(GO:0033131) positive regulation of glucokinase activity(GO:0033133) |
| 0.0 | 0.2 | GO:0033601 | positive regulation of mammary gland epithelial cell proliferation(GO:0033601) |
| 0.0 | 0.0 | GO:0072004 | pronephric field specification(GO:0039003) pattern specification involved in pronephros development(GO:0039017) kidney field specification(GO:0072004) regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072304) negative regulation of mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:0072305) apoptotic process involved in metanephric collecting duct development(GO:1900204) apoptotic process involved in metanephric nephron tubule development(GO:1900205) regulation of apoptotic process involved in metanephric collecting duct development(GO:1900214) negative regulation of apoptotic process involved in metanephric collecting duct development(GO:1900215) regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900217) negative regulation of apoptotic process involved in metanephric nephron tubule development(GO:1900218) mesenchymal cell apoptotic process involved in metanephric nephron morphogenesis(GO:1901147) regulation of metanephric DCT cell differentiation(GO:2000592) positive regulation of metanephric DCT cell differentiation(GO:2000594) |
| 0.0 | 0.2 | GO:0098915 | membrane repolarization during ventricular cardiac muscle cell action potential(GO:0098915) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.0 | GO:0010813 | neuropeptide catabolic process(GO:0010813) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0044804 | nucleophagy(GO:0044804) |
| 0.0 | 0.1 | GO:0050915 | sensory perception of sour taste(GO:0050915) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.2 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.1 | GO:0090063 | positive regulation of microtubule nucleation(GO:0090063) |
| 0.0 | 0.3 | GO:0098780 | response to mitochondrial depolarisation(GO:0098780) |
| 0.0 | 0.1 | GO:1901382 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.2 | GO:0030206 | chondroitin sulfate biosynthetic process(GO:0030206) |
| 0.0 | 0.1 | GO:0061072 | lens induction in camera-type eye(GO:0060235) iris morphogenesis(GO:0061072) |
| 0.0 | 0.1 | GO:2000327 | positive regulation of ligand-dependent nuclear receptor transcription coactivator activity(GO:2000327) |
| 0.0 | 0.2 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.2 | GO:0000478 | endonucleolytic cleavage involved in rRNA processing(GO:0000478) endonucleolytic cleavage of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000479) |
| 0.0 | 0.2 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.0 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) |
| 0.0 | 0.2 | GO:0009396 | folic acid-containing compound biosynthetic process(GO:0009396) |
| 0.0 | 0.3 | GO:0010257 | NADH dehydrogenase complex assembly(GO:0010257) mitochondrial respiratory chain complex I assembly(GO:0032981) mitochondrial respiratory chain complex I biogenesis(GO:0097031) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.2 | GO:0007097 | nuclear migration(GO:0007097) |
| 0.0 | 0.0 | GO:0010593 | negative regulation of lamellipodium assembly(GO:0010593) |
| 0.0 | 0.1 | GO:0030948 | negative regulation of vascular endothelial growth factor receptor signaling pathway(GO:0030948) |
| 0.0 | 0.1 | GO:0032342 | aldosterone biosynthetic process(GO:0032342) |
| 0.0 | 0.1 | GO:0060040 | retinal bipolar neuron differentiation(GO:0060040) |
| 0.0 | 0.1 | GO:0060708 | negative regulation of Schwann cell proliferation(GO:0010626) spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0051344 | negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.1 | GO:0044691 | tooth eruption(GO:0044691) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.1 | GO:0001923 | B-1 B cell differentiation(GO:0001923) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:1904153 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.2 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0070827 | chromatin maintenance(GO:0070827) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0038037 | G-protein coupled receptor dimeric complex(GO:0038037) G-protein coupled receptor complex(GO:0097648) |
| 0.1 | 0.4 | GO:0071203 | WASH complex(GO:0071203) |
| 0.1 | 0.2 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.1 | 0.3 | GO:0070033 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin II complex(GO:0070033) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0090534 | longitudinal sarcoplasmic reticulum(GO:0014801) calcium ion-transporting ATPase complex(GO:0090534) |
| 0.0 | 0.2 | GO:0005869 | dynactin complex(GO:0005869) |
| 0.0 | 0.3 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.3 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.4 | GO:0044224 | juxtaparanode region of axon(GO:0044224) |
| 0.0 | 0.2 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.0 | GO:0070032 | synaptobrevin 2-SNAP-25-syntaxin-1a-complexin I complex(GO:0070032) |
| 0.0 | 0.5 | GO:0030285 | integral component of synaptic vesicle membrane(GO:0030285) intrinsic component of synaptic vesicle membrane(GO:0098563) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0032280 | symmetric synapse(GO:0032280) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.2 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0030128 | clathrin coat of endocytic vesicle(GO:0030128) |
| 0.0 | 0.4 | GO:0032809 | neuronal cell body membrane(GO:0032809) |
| 0.0 | 0.1 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0036513 | Derlin-1 retrotranslocation complex(GO:0036513) |
| 0.0 | 0.8 | GO:0005788 | endoplasmic reticulum lumen(GO:0005788) |
| 0.0 | 0.1 | GO:0033093 | Weibel-Palade body(GO:0033093) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0017168 | 5-oxoprolinase (ATP-hydrolyzing) activity(GO:0017168) |
| 0.1 | 0.6 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.3 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 1.0 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.2 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.1 | 0.2 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.1 | 0.4 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.4 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0050265 | RNA uridylyltransferase activity(GO:0050265) |
| 0.0 | 1.1 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0051998 | carboxyl-O-methyltransferase activity(GO:0010340) protein carboxyl O-methyltransferase activity(GO:0051998) |
| 0.0 | 0.2 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0008889 | glycerophosphodiester phosphodiesterase activity(GO:0008889) |
| 0.0 | 0.1 | GO:1990763 | arrestin family protein binding(GO:1990763) |
| 0.0 | 0.1 | GO:0004507 | steroid 11-beta-monooxygenase activity(GO:0004507) corticosterone 18-monooxygenase activity(GO:0047783) |
| 0.0 | 0.2 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.4 | GO:0010314 | phosphatidylinositol-5-phosphate binding(GO:0010314) |
| 0.0 | 0.2 | GO:0034481 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) chondroitin sulfotransferase activity(GO:0034481) |
| 0.0 | 0.3 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0001155 | TFIIIA-class transcription factor binding(GO:0001155) |
| 0.0 | 0.1 | GO:0003726 | double-stranded RNA adenosine deaminase activity(GO:0003726) |
| 0.0 | 0.1 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.1 | GO:0001156 | TFIIIC-class transcription factor binding(GO:0001156) |
| 0.0 | 0.1 | GO:0005042 | netrin receptor activity(GO:0005042) |
| 0.0 | 0.3 | GO:0098988 | G-protein coupled glutamate receptor activity(GO:0098988) |
| 0.0 | 0.5 | GO:0005159 | insulin-like growth factor receptor binding(GO:0005159) |
| 0.0 | 0.1 | GO:0051731 | polynucleotide 5'-hydroxyl-kinase activity(GO:0051731) |
| 0.0 | 0.1 | GO:0000991 | transcription factor activity, core RNA polymerase II binding(GO:0000991) |
| 0.0 | 0.2 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.2 | GO:0034511 | U3 snoRNA binding(GO:0034511) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.0 | 0.2 | GO:0051766 | inositol trisphosphate kinase activity(GO:0051766) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.2 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.0 | GO:0004307 | ethanolaminephosphotransferase activity(GO:0004307) |
| 0.0 | 0.4 | GO:0004623 | phospholipase A2 activity(GO:0004623) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0016493 | C-C chemokine receptor activity(GO:0016493) |
| 0.0 | 0.4 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.2 | GO:0031434 | mitogen-activated protein kinase kinase binding(GO:0031434) |
| 0.0 | 0.2 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.1 | GO:0050692 | DBD domain binding(GO:0050692) |
Gene overrepresentation in curated gene sets: canonical pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.8 | ST TUMOR NECROSIS FACTOR PATHWAY | Tumor Necrosis Factor Pathway. |
| 0.0 | 0.2 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.7 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 0.2 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.7 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.0 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.3 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.0 | 0.1 | REACTOME INFLUENZA VIRAL RNA TRANSCRIPTION AND REPLICATION | Genes involved in Influenza Viral RNA Transcription and Replication |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.2 | REACTOME ACETYLCHOLINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Acetylcholine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 0.2 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |