Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
Results for Pax4
Z-value: 0.37
Transcription factors associated with Pax4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Pax4
|
ENSRNOG00000008020 | paired box 4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Pax4 | rn6_v1_chr4_-_55740627_55740627 | -0.35 | 5.6e-01 | Click! |
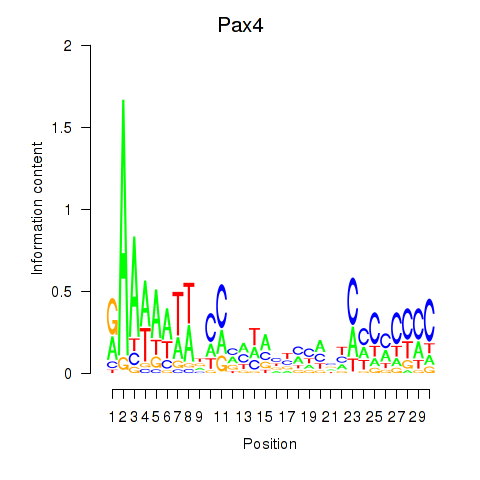
Activity profile of Pax4 motif
Sorted Z-values of Pax4 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_+_79823945 | 0.15 |
ENSRNOT00000014484
|
Celf1
|
CUGBP, Elav-like family member 1 |
| chr9_+_20765296 | 0.09 |
ENSRNOT00000016291
|
Cd2ap
|
CD2-associated protein |
| chr13_-_45318822 | 0.09 |
ENSRNOT00000091514
ENSRNOT00000005143 |
Cxcr4
|
C-X-C motif chemokine receptor 4 |
| chr6_-_108796124 | 0.08 |
ENSRNOT00000086545
|
Arel1
|
apoptosis resistant E3 ubiquitin protein ligase 1 |
| chr16_-_9709347 | 0.07 |
ENSRNOT00000083933
|
Mapk8
|
mitogen-activated protein kinase 8 |
| chr20_+_13940877 | 0.07 |
ENSRNOT00000093587
|
Cabin1
|
calcineurin binding protein 1 |
| chr4_-_152883210 | 0.07 |
ENSRNOT00000056200
|
Ccdc77
|
coiled-coil domain containing 77 |
| chrX_+_128416722 | 0.07 |
ENSRNOT00000009336
ENSRNOT00000085110 |
Xiap
|
X-linked inhibitor of apoptosis |
| chr3_-_148932878 | 0.07 |
ENSRNOT00000013881
|
Nol4l
|
nucleolar protein 4-like |
| chr18_-_74485139 | 0.06 |
ENSRNOT00000022598
|
Slc14a1
|
solute carrier family 14 member 1 |
| chr18_-_57245666 | 0.06 |
ENSRNOT00000080365
|
Ablim3
|
actin binding LIM protein family, member 3 |
| chr3_-_71845232 | 0.06 |
ENSRNOT00000078645
|
Calcrl
|
calcitonin receptor like receptor |
| chr13_-_80775230 | 0.06 |
ENSRNOT00000091389
ENSRNOT00000004762 |
Fmo2
|
flavin containing monooxygenase 2 |
| chr8_+_70317586 | 0.05 |
ENSRNOT00000016260
|
Dennd4a
|
DENN domain containing 4A |
| chr2_-_22798214 | 0.05 |
ENSRNOT00000016135
|
Papd4
|
poly(A) RNA polymerase D4, non-canonical |
| chr3_-_113438801 | 0.05 |
ENSRNOT00000091665
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chrX_-_152642531 | 0.05 |
ENSRNOT00000085037
|
Gabra3
|
gamma-aminobutyric acid type A receptor alpha3 subunit |
| chr14_-_112946204 | 0.05 |
ENSRNOT00000056813
|
LOC103690141
|
coiled-coil domain-containing protein 85A-like |
| chrX_-_70428364 | 0.05 |
ENSRNOT00000045907
|
P2ry4
|
pyrimidinergic receptor P2Y4 |
| chr14_+_8182383 | 0.05 |
ENSRNOT00000092719
|
Mapk10
|
mitogen activated protein kinase 10 |
| chr10_+_90984227 | 0.05 |
ENSRNOT00000003890
ENSRNOT00000093707 |
Ccdc103
|
coiled-coil domain containing 103 |
| chr1_+_264309214 | 0.04 |
ENSRNOT00000019070
|
Hif1an
|
hypoxia-inducible factor 1, alpha subunit inhibitor |
| chr3_-_66417741 | 0.04 |
ENSRNOT00000007662
|
Neurod1
|
neuronal differentiation 1 |
| chr3_-_162579201 | 0.04 |
ENSRNOT00000068328
|
Zmynd8
|
zinc finger, MYND-type containing 8 |
| chr1_+_105285419 | 0.04 |
ENSRNOT00000089693
|
Slc6a5
|
solute carrier family 6 member 5 |
| chrM_+_11736 | 0.04 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr1_+_252894663 | 0.04 |
ENSRNOT00000054757
|
Ifit2
|
interferon-induced protein with tetratricopeptide repeats 2 |
| chr14_+_39663421 | 0.04 |
ENSRNOT00000003197
|
Gabra2
|
gamma-aminobutyric acid type A receptor alpha2 subunit |
| chr1_+_81779380 | 0.04 |
ENSRNOT00000065865
ENSRNOT00000080143 ENSRNOT00000089592 ENSRNOT00000080840 |
Arhgef1
|
Rho guanine nucleotide exchange factor 1 |
| chr6_-_50941248 | 0.04 |
ENSRNOT00000084533
|
Dus4l
|
dihydrouridine synthase 4-like |
| chr7_+_97559841 | 0.03 |
ENSRNOT00000007326
|
Zhx2
|
zinc fingers and homeoboxes 2 |
| chr5_+_6373583 | 0.03 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr7_-_140437467 | 0.03 |
ENSRNOT00000087181
|
Fkbp11
|
FK506 binding protein 11 |
| chr8_+_12823155 | 0.03 |
ENSRNOT00000011008
|
Sesn3
|
sestrin 3 |
| chr3_-_51297852 | 0.03 |
ENSRNOT00000001607
|
Cobll1
|
cordon-bleu WH2 repeat protein-like 1 |
| chr7_+_117605050 | 0.03 |
ENSRNOT00000047380
|
Slc52a2
|
solute carrier family 52 member 2 |
| chr7_-_120295618 | 0.03 |
ENSRNOT00000014413
|
Ankrd54
|
ankyrin repeat domain 54 |
| chrM_+_9870 | 0.03 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr14_-_103321270 | 0.03 |
ENSRNOT00000006157
|
Meis1
|
Meis homeobox 1 |
| chr15_-_42947656 | 0.03 |
ENSRNOT00000030007
|
Ptk2b
|
protein tyrosine kinase 2 beta |
| chr7_-_117605141 | 0.03 |
ENSRNOT00000035199
|
Fbxl6
|
F-box and leucine-rich repeat protein 6 |
| chr12_-_37411525 | 0.03 |
ENSRNOT00000087698
|
Tctn2
|
tectonic family member 2 |
| chr14_+_99919485 | 0.02 |
ENSRNOT00000006087
|
Egfr
|
epidermal growth factor receptor |
| chr8_-_53901358 | 0.02 |
ENSRNOT00000047666
ENSRNOT00000042281 |
Ncam1
|
neural cell adhesion molecule 1 |
| chr1_+_116604550 | 0.02 |
ENSRNOT00000083587
|
Ube3a
|
ubiquitin protein ligase E3A |
| chr20_-_6257604 | 0.02 |
ENSRNOT00000092489
|
Stk38
|
serine/threonine kinase 38 |
| chr3_-_92749121 | 0.02 |
ENSRNOT00000008760
|
Cd44
|
CD44 molecule (Indian blood group) |
| chr20_-_1878126 | 0.02 |
ENSRNOT00000000995
|
Ubd
|
ubiquitin D |
| chr12_-_11176106 | 0.02 |
ENSRNOT00000090073
|
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr1_-_221370322 | 0.02 |
ENSRNOT00000028431
|
Capn1
|
calpain 1 |
| chr12_-_11175917 | 0.02 |
ENSRNOT00000079573
|
Zkscan5
|
zinc finger with KRAB and SCAN domains 5 |
| chr15_+_43298794 | 0.02 |
ENSRNOT00000012736
|
Adra1a
|
adrenoceptor alpha 1A |
| chr15_-_6587367 | 0.02 |
ENSRNOT00000038449
|
Zfp385d
|
zinc finger protein 385D |
| chrX_-_116792707 | 0.02 |
ENSRNOT00000049482
|
Amot
|
angiomotin |
| chr20_-_4401610 | 0.02 |
ENSRNOT00000091468
|
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr3_+_94530586 | 0.02 |
ENSRNOT00000067860
|
Cstf3
|
cleavage stimulation factor subunit 3 |
| chr7_+_37018459 | 0.02 |
ENSRNOT00000087297
|
Eea1
|
early endosome antigen 1 |
| chr7_-_101140308 | 0.02 |
ENSRNOT00000006279
|
Fam84b
|
family with sequence similarity 84, member B |
| chr5_+_148781222 | 0.02 |
ENSRNOT00000015831
|
Pum1
|
pumilio RNA-binding family member 1 |
| chr9_-_10471009 | 0.02 |
ENSRNOT00000072868
|
Safb
|
scaffold attachment factor B |
| chr11_-_57993548 | 0.01 |
ENSRNOT00000002957
|
Nectin3
|
nectin cell adhesion molecule 3 |
| chr12_-_44999074 | 0.01 |
ENSRNOT00000079177
ENSRNOT00000038293 |
Vsig10
|
V-set and immunoglobulin domain containing 10 |
| chr7_-_114573900 | 0.01 |
ENSRNOT00000011219
|
Ptk2
|
protein tyrosine kinase 2 |
| chr1_-_248376750 | 0.01 |
ENSRNOT00000089073
|
Gldc
|
glycine decarboxylase |
| chr7_-_119689938 | 0.01 |
ENSRNOT00000000200
|
Tmprss6
|
transmembrane protease, serine 6 |
| chr18_-_81682206 | 0.01 |
ENSRNOT00000058219
|
LOC100359752
|
hypothetical protein LOC100359752 |
| chr8_-_67040005 | 0.01 |
ENSRNOT00000038641
|
Glce
|
glucuronic acid epimerase |
| chr1_+_80092403 | 0.01 |
ENSRNOT00000078336
|
Eml2
|
echinoderm microtubule associated protein like 2 |
| chr20_-_44255064 | 0.01 |
ENSRNOT00000000735
|
Wisp3
|
WNT1 inducible signaling pathway protein 3 |
| chr20_-_5076539 | 0.01 |
ENSRNOT00000076253
ENSRNOT00000037613 |
Ly6g6f
|
lymphocyte antigen 6 complex, locus G6F |
| chr6_+_28235695 | 0.01 |
ENSRNOT00000047210
|
Dnmt3a
|
DNA methyltransferase 3 alpha |
| chr13_+_49005405 | 0.01 |
ENSRNOT00000092560
ENSRNOT00000076457 |
Lemd1
|
LEM domain containing 1 |
| chr8_+_107508351 | 0.01 |
ENSRNOT00000081594
|
Cep70
|
centrosomal protein 70 |
| chr4_+_110699557 | 0.01 |
ENSRNOT00000030588
ENSRNOT00000092261 |
Lrrtm4
|
leucine rich repeat transmembrane neuronal 4 |
| chrX_+_105537602 | 0.01 |
ENSRNOT00000029833
|
Armcx1
|
armadillo repeat containing, X-linked 1 |
| chr12_+_40244081 | 0.01 |
ENSRNOT00000030583
|
Sh2b3
|
SH2B adaptor protein 3 |
| chr4_-_158704170 | 0.01 |
ENSRNOT00000082009
|
Ntf3
|
neurotrophin 3 |
| chrM_+_3904 | 0.01 |
ENSRNOT00000040993
|
Mt-nd2
|
mitochondrially encoded NADH dehydrogenase 2 |
| chr6_-_103313074 | 0.01 |
ENSRNOT00000083677
|
Zfp36l1
|
zinc finger protein 36, C3H type-like 1 |
| chr18_+_60392376 | 0.00 |
ENSRNOT00000023890
|
Nedd4l
|
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr13_-_53108713 | 0.00 |
ENSRNOT00000035404
|
RGD1311892
|
similar to hypothetical protein FLJ10901 |
| chr13_+_90301006 | 0.00 |
ENSRNOT00000029315
|
Slamf6
|
SLAM family member 6 |
| chr1_+_273854248 | 0.00 |
ENSRNOT00000044827
ENSRNOT00000017600 ENSRNOT00000086496 |
Add3
|
adducin 3 |
| chr16_-_62164135 | 0.00 |
ENSRNOT00000058872
|
Gtf2e2
|
general transcription factor IIE subunit 2 |
| chr18_+_30527705 | 0.00 |
ENSRNOT00000027168
|
Pcdhb14
|
protocadherin beta 14 |
| chr10_-_25845634 | 0.00 |
ENSRNOT00000004269
|
Mat2b
|
methionine adenosyltransferase 2B |
| chr5_+_117583502 | 0.00 |
ENSRNOT00000010933
|
Usp1
|
ubiquitin specific peptidase 1 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Pax4
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:1990478 | positive regulation of vascular wound healing(GO:0035470) response to ultrasound(GO:1990478) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0097039 | protein linear polyubiquitination(GO:0097039) |
| 0.0 | 0.0 | GO:0007172 | signal complex assembly(GO:0007172) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0001605 | adrenomedullin receptor activity(GO:0001605) |
| 0.0 | 0.1 | GO:0000827 | inositol-1,3,4,5,6-pentakisphosphate kinase activity(GO:0000827) |
| 0.0 | 0.1 | GO:0015065 | uridine nucleotide receptor activity(GO:0015065) G-protein coupled pyrimidinergic nucleotide receptor activity(GO:0071553) |
| 0.0 | 0.1 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.0 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.1 | GO:0004705 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |