Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
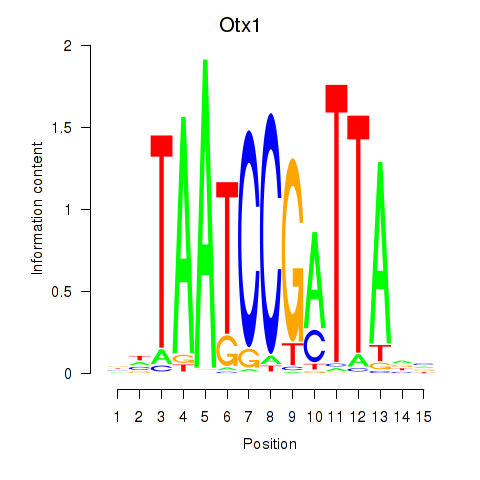
Results for Otx1
Z-value: 0.30
Transcription factors associated with Otx1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Otx1
|
ENSRNOG00000059469 | orthodenticle homeobox 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Otx1 | rn6_v1_chr14_-_106864892_106864892 | 0.74 | 1.5e-01 | Click! |
Activity profile of Otx1 motif
Sorted Z-values of Otx1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_27047382 | 0.12 |
ENSRNOT00000027691
ENSRNOT00000090264 |
Apc
|
APC, WNT signaling pathway regulator |
| chr2_-_265300868 | 0.11 |
ENSRNOT00000066024
ENSRNOT00000016073 ENSRNOT00000033502 |
Lrrc7
|
leucine rich repeat containing 7 |
| chr19_+_26066127 | 0.11 |
ENSRNOT00000064289
|
Rtbdn
|
retbindin |
| chr10_+_56381813 | 0.10 |
ENSRNOT00000019687
|
Zbtb4
|
zinc finger and BTB domain containing 4 |
| chr1_+_27476375 | 0.10 |
ENSRNOT00000047224
ENSRNOT00000075427 |
LOC102551716
|
sodium/potassium-transporting ATPase subunit beta-1-interacting protein 2-like |
| chr3_-_162059524 | 0.08 |
ENSRNOT00000033873
|
Zfp334
|
zinc finger protein 334 |
| chr7_+_70946228 | 0.08 |
ENSRNOT00000039306
|
Stat6
|
signal transducer and activator of transcription 6 |
| chrM_+_11736 | 0.08 |
ENSRNOT00000048767
|
Mt-nd5
|
mitochondrially encoded NADH dehydrogenase 5 |
| chr13_-_110784209 | 0.07 |
ENSRNOT00000071698
|
Traf5
|
TNF receptor-associated factor 5 |
| chr3_+_121596791 | 0.07 |
ENSRNOT00000024694
|
Ttl
|
tubulin tyrosine ligase |
| chr2_+_66940057 | 0.07 |
ENSRNOT00000043050
|
Cdh9
|
cadherin 9 |
| chr14_-_71973419 | 0.07 |
ENSRNOT00000079895
|
Cc2d2a
|
coiled-coil and C2 domain containing 2A |
| chr11_+_57207656 | 0.07 |
ENSRNOT00000038207
ENSRNOT00000085754 |
Plcxd2
|
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr14_-_46153212 | 0.06 |
ENSRNOT00000079269
|
Nwd2
|
NACHT and WD repeat domain containing 2 |
| chr2_+_196594303 | 0.06 |
ENSRNOT00000064442
ENSRNOT00000044738 |
Arnt
|
aryl hydrocarbon receptor nuclear translocator |
| chr12_-_10391270 | 0.06 |
ENSRNOT00000092340
|
Wasf3
|
WAS protein family, member 3 |
| chr1_+_65522118 | 0.06 |
ENSRNOT00000058563
|
Mzf1
|
myeloid zinc finger 1 |
| chrX_+_984798 | 0.06 |
ENSRNOT00000073016
|
Zfp182
|
zinc finger protein 182 |
| chr15_+_19547871 | 0.06 |
ENSRNOT00000036235
|
Gpr137c
|
G protein-coupled receptor 137C |
| chrX_+_128416722 | 0.06 |
ENSRNOT00000009336
ENSRNOT00000085110 |
Xiap
|
X-linked inhibitor of apoptosis |
| chr4_-_183293999 | 0.06 |
ENSRNOT00000079695
|
Ipo8
|
importin 8 |
| chr18_+_15856801 | 0.06 |
ENSRNOT00000071548
|
Zfp397
|
zinc finger protein 397 |
| chr10_-_62814520 | 0.05 |
ENSRNOT00000019410
|
Ssh2
|
slingshot protein phosphatase 2 |
| chr10_+_57218087 | 0.05 |
ENSRNOT00000089853
|
Mink1
|
misshapen-like kinase 1 |
| chrX_+_22302703 | 0.05 |
ENSRNOT00000078617
ENSRNOT00000086952 |
Kdm5c
|
lysine demethylase 5C |
| chr16_+_72268943 | 0.05 |
ENSRNOT00000032116
|
Ido2
|
indoleamine 2,3-dioxygenase 2 |
| chr4_+_9981958 | 0.05 |
ENSRNOT00000015322
|
Napepld
|
N-acyl phosphatidylethanolamine phospholipase D |
| chr1_+_224998172 | 0.05 |
ENSRNOT00000026321
|
Zbtb3
|
zinc finger and BTB domain containing 3 |
| chr17_-_27602934 | 0.05 |
ENSRNOT00000079298
|
Rreb1
|
ras responsive element binding protein 1 |
| chrX_+_22302485 | 0.05 |
ENSRNOT00000082902
|
Kdm5c
|
lysine demethylase 5C |
| chr16_+_10727571 | 0.05 |
ENSRNOT00000084422
|
Mmrn2
|
multimerin 2 |
| chr17_-_28191436 | 0.05 |
ENSRNOT00000000149
|
Ly86
|
lymphocyte antigen 86 |
| chr3_-_21684132 | 0.05 |
ENSRNOT00000012364
|
Zbtb6
|
zinc finger and BTB domain containing 6 |
| chr2_+_113984824 | 0.04 |
ENSRNOT00000086399
|
Tnik
|
TRAF2 and NCK interacting kinase |
| chr18_+_30496318 | 0.04 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr6_-_114476723 | 0.04 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chrX_+_68771100 | 0.04 |
ENSRNOT00000043872
|
Stard8
|
StAR-related lipid transfer domain containing 8 |
| chr4_+_87026530 | 0.04 |
ENSRNOT00000018425
|
Avl9
|
AVL9 cell migration associated |
| chr14_+_37919233 | 0.04 |
ENSRNOT00000003125
|
Tec
|
tec protein tyrosine kinase |
| chr18_+_30909490 | 0.04 |
ENSRNOT00000026967
|
Pcdhgb8
|
protocadherin gamma subfamily B, 8 |
| chr2_+_93520734 | 0.04 |
ENSRNOT00000083909
|
Snx16
|
sorting nexin 16 |
| chr7_+_97760134 | 0.04 |
ENSRNOT00000086792
|
Tbc1d31
|
TBC1 domain family, member 31 |
| chr12_+_31536951 | 0.04 |
ENSRNOT00000086078
|
Rimbp2
|
RIMS binding protein 2 |
| chr9_+_49837868 | 0.04 |
ENSRNOT00000021999
|
Gpr45
|
G protein-coupled receptor 45 |
| chr20_+_6211420 | 0.03 |
ENSRNOT00000000624
|
Kctd20
|
potassium channel tetramerization domain containing 20 |
| chr6_-_91581262 | 0.03 |
ENSRNOT00000034507
|
AABR07064716.1
|
|
| chr15_-_33250546 | 0.03 |
ENSRNOT00000017857
|
RGD1565222
|
similar to RIKEN cDNA 4931414P19 |
| chr6_-_91676344 | 0.03 |
ENSRNOT00000083872
|
Nemf
|
nuclear export mediator factor |
| chr1_-_275882444 | 0.03 |
ENSRNOT00000083215
|
Gpam
|
glycerol-3-phosphate acyltransferase, mitochondrial |
| chr7_-_129919946 | 0.03 |
ENSRNOT00000079976
|
AABR07058658.1
|
|
| chr1_+_239412884 | 0.03 |
ENSRNOT00000067384
|
Tmem2
|
transmembrane protein 2 |
| chr12_-_12945192 | 0.03 |
ENSRNOT00000082529
|
Cyth3
|
cytohesin 3 |
| chr1_-_80594136 | 0.03 |
ENSRNOT00000024800
|
Apoc2
|
apolipoprotein C2 |
| chr14_-_16903242 | 0.03 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr1_-_87777668 | 0.03 |
ENSRNOT00000037288
|
AABR07002870.1
|
|
| chr4_-_117589464 | 0.03 |
ENSRNOT00000021167
|
Nat8f1
|
N-acetyltransferase 8 (GCN5-related) family member 1 |
| chr3_+_130008885 | 0.03 |
ENSRNOT00000070972
|
Slx4ip
|
SLX4 interacting protein |
| chr7_-_106753592 | 0.03 |
ENSRNOT00000006930
|
Kcnq3
|
potassium voltage-gated channel subfamily Q member 3 |
| chr17_+_44758460 | 0.03 |
ENSRNOT00000089436
|
Hist1h2an
|
histone cluster 1, H2an |
| chr9_+_12114977 | 0.03 |
ENSRNOT00000073673
|
AABR07066648.1
|
|
| chr13_-_39643361 | 0.03 |
ENSRNOT00000003527
|
Dpp10
|
dipeptidylpeptidase 10 |
| chr3_-_152259156 | 0.03 |
ENSRNOT00000065729
|
LOC100910882
|
RNA-binding protein 39-like |
| chrX_-_56765893 | 0.03 |
ENSRNOT00000076283
|
Il1rapl1
|
interleukin 1 receptor accessory protein-like 1 |
| chr5_+_139385429 | 0.03 |
ENSRNOT00000078622
|
Scmh1
|
sex comb on midleg homolog 1 (Drosophila) |
| chrX_+_123751089 | 0.03 |
ENSRNOT00000092384
|
Nkap
|
NFKB activating protein |
| chr2_-_106703401 | 0.02 |
ENSRNOT00000043933
|
AABR07009638.1
|
|
| chr18_+_40883224 | 0.02 |
ENSRNOT00000039733
|
Lvrn
|
laeverin |
| chr19_+_37252843 | 0.02 |
ENSRNOT00000021145
|
E2f4
|
E2F transcription factor 4 |
| chr3_-_151724654 | 0.02 |
ENSRNOT00000026964
|
Rbm39
|
RNA binding motif protein 39 |
| chr13_-_25652473 | 0.02 |
ENSRNOT00000020752
|
Pign
|
phosphatidylinositol glycan anchor biosynthesis, class N |
| chr14_+_7630482 | 0.02 |
ENSRNOT00000091581
|
LOC498330
|
similar to hypothetical protein MGC26744 |
| chr2_+_102685513 | 0.02 |
ENSRNOT00000033940
|
Bhlhe22
|
basic helix-loop-helix family, member e22 |
| chr15_-_52029816 | 0.02 |
ENSRNOT00000013067
|
Slc39a14
|
solute carrier family 39 member 14 |
| chr1_+_69853553 | 0.02 |
ENSRNOT00000042086
|
Zfp954
|
zinc finger protein 954 |
| chr7_+_123963381 | 0.02 |
ENSRNOT00000036586
|
Serhl2
|
serine hydrolase-like 2 |
| chr1_-_90035522 | 0.02 |
ENSRNOT00000028672
|
Uba2
|
ubiquitin-like modifier activating enzyme 2 |
| chr1_-_127292090 | 0.02 |
ENSRNOT00000016959
|
Lrrk1
|
leucine-rich repeat kinase 1 |
| chr1_-_214414763 | 0.02 |
ENSRNOT00000025240
ENSRNOT00000077776 |
LOC100911440
|
mitochondrial glutamate carrier 1-like |
| chr9_-_9865920 | 0.02 |
ENSRNOT00000071068
|
Brcc3
|
BRCA1/BRCA2-containing complex, subunit 3 |
| chr8_-_116855062 | 0.02 |
ENSRNOT00000050344
|
Rnf123
|
ring finger protein 123 |
| chr8_+_82038967 | 0.02 |
ENSRNOT00000079535
|
Myo5a
|
myosin VA |
| chr9_-_65442257 | 0.02 |
ENSRNOT00000037660
|
Fam126b
|
family with sequence similarity 126, member B |
| chr2_-_30791221 | 0.02 |
ENSRNOT00000082846
|
Ccnb1
|
cyclin B1 |
| chr14_+_45033309 | 0.02 |
ENSRNOT00000002933
|
Tlr6
|
toll-like receptor 6 |
| chr8_-_57830504 | 0.02 |
ENSRNOT00000017683
|
Ddx10
|
DEAD-box helicase 10 |
| chr11_+_43194348 | 0.02 |
ENSRNOT00000075303
|
Olr1532
|
olfactory receptor 1532 |
| chr5_+_128083118 | 0.02 |
ENSRNOT00000079161
|
Zcchc11
|
zinc finger CCHC-type containing 11 |
| chr8_+_93439648 | 0.02 |
ENSRNOT00000043008
|
LOC100359563
|
ribosomal protein S20-like |
| chr8_+_117280705 | 0.02 |
ENSRNOT00000085038
|
Usp19
|
ubiquitin specific peptidase 19 |
| chr1_-_201942344 | 0.02 |
ENSRNOT00000027956
|
RGD1305014
|
similar to RIKEN cDNA 2310057M21 |
| chr3_-_57607683 | 0.01 |
ENSRNOT00000093222
ENSRNOT00000058524 |
Mettl8
|
methyltransferase like 8 |
| chr3_+_111826297 | 0.01 |
ENSRNOT00000081897
|
Pla2g4b
|
phospholipase A2 group IVB |
| chr10_+_45258887 | 0.01 |
ENSRNOT00000048642
|
Btnl10
|
butyrophilin like 10 |
| chr3_+_67870111 | 0.01 |
ENSRNOT00000012628
|
Nup35
|
nucleoporin 35 |
| chr12_-_24724997 | 0.01 |
ENSRNOT00000025560
|
Abhd11
|
abhydrolase domain containing 11 |
| chr1_-_217620420 | 0.01 |
ENSRNOT00000092783
|
Cttn
|
cortactin |
| chr10_-_90569575 | 0.01 |
ENSRNOT00000074987
|
AABR07030494.1
|
|
| chr13_-_83202864 | 0.01 |
ENSRNOT00000003976
|
Xcl1
|
X-C motif chemokine ligand 1 |
| chr18_+_36829062 | 0.01 |
ENSRNOT00000025467
ENSRNOT00000086979 |
Tcerg1
|
transcription elongation regulator 1 |
| chr7_-_70467915 | 0.01 |
ENSRNOT00000088995
|
Slc26a10
|
solute carrier family 26, member 10 |
| chr4_+_145195070 | 0.01 |
ENSRNOT00000010723
|
Mtmr14
|
myotubularin related protein 14 |
| chr11_+_64522130 | 0.01 |
ENSRNOT00000004315
|
Upk1b
|
uroplakin 1B |
| chr5_+_6373583 | 0.01 |
ENSRNOT00000084749
|
AABR07046778.1
|
|
| chr1_-_155955173 | 0.01 |
ENSRNOT00000079345
|
AABR07004776.1
|
|
| chr8_-_104995725 | 0.01 |
ENSRNOT00000037120
|
Slc25a36
|
solute carrier family 25 member 36 |
| chr5_+_156396695 | 0.01 |
ENSRNOT00000067631
|
Eif4g3
|
eukaryotic translation initiation factor 4 gamma, 3 |
| chr6_+_26836216 | 0.01 |
ENSRNOT00000077903
ENSRNOT00000011373 |
Ost4
|
oligosaccharyltransferase complex subunit 4, non-catalytic |
| chr1_+_80954858 | 0.01 |
ENSRNOT00000030440
|
Zfp112
|
zinc finger protein 112 |
| chr9_-_71798265 | 0.01 |
ENSRNOT00000043766
|
Crygb
|
crystallin, gamma B |
| chr7_+_121350965 | 0.01 |
ENSRNOT00000092614
|
Tab1
|
TGF-beta activated kinase 1/MAP3K7 binding protein 1 |
| chr5_+_61474000 | 0.01 |
ENSRNOT00000013930
|
Ccdc180
|
coiled-coil domain containing 180 |
| chr7_-_111722964 | 0.01 |
ENSRNOT00000041802
|
Rps19l1
|
ribosomal protein S19-like 1 |
| chr14_+_36071376 | 0.01 |
ENSRNOT00000082183
|
Lnx1
|
ligand of numb-protein X 1 |
| chr1_+_260153645 | 0.01 |
ENSRNOT00000054717
|
Zfp518a
|
zinc finger protein 518A |
| chr7_-_3229167 | 0.01 |
ENSRNOT00000008929
|
Tmem198b
|
transmembrane protein 198b |
| chr3_+_118430511 | 0.01 |
ENSRNOT00000012966
|
Dtwd1
|
DTW domain containing 1 |
| chr11_+_31806618 | 0.01 |
ENSRNOT00000043410
ENSRNOT00000002769 |
Son
|
Son DNA binding protein |
| chr8_-_62410338 | 0.01 |
ENSRNOT00000026358
|
Csk
|
c-src tyrosine kinase |
| chrM_+_9870 | 0.01 |
ENSRNOT00000044582
|
Mt-nd4l
|
mitochondrially encoded NADH 4L dehydrogenase |
| chr7_-_124020574 | 0.01 |
ENSRNOT00000055988
|
Poldip3
|
DNA polymerase delta interacting protein 3 |
| chr7_-_62162453 | 0.01 |
ENSRNOT00000010720
|
Cand1
|
cullin-associated and neddylation-dissociated 1 |
| chr3_+_80362858 | 0.01 |
ENSRNOT00000021353
ENSRNOT00000086242 |
Lrp4
|
LDL receptor related protein 4 |
| chr3_-_93693690 | 0.01 |
ENSRNOT00000086745
|
Nat10
|
N-acetyltransferase 10 |
| chr12_-_8418966 | 0.01 |
ENSRNOT00000085869
|
Mtus2
|
microtubule associated tumor suppressor candidate 2 |
| chr1_-_166919302 | 0.01 |
ENSRNOT00000026925
|
Folr2
|
folate receptor beta |
| chr8_+_116336441 | 0.01 |
ENSRNOT00000090123
ENSRNOT00000021590 |
Nat6
Hyal3
|
N-acetyltransferase 6 hyaluronoglucosaminidase 3 |
| chr17_-_19703681 | 0.01 |
ENSRNOT00000024021
|
Mylip
|
myosin regulatory light chain interacting protein |
| chr20_-_14573519 | 0.01 |
ENSRNOT00000001772
|
Rab36
|
RAB36, member RAS oncogene family |
| chr6_+_92864123 | 0.00 |
ENSRNOT00000087540
|
Tmx1
|
thioredoxin-related transmembrane protein 1 |
| chr4_+_144985880 | 0.00 |
ENSRNOT00000009464
|
Thumpd3
|
THUMP domain containing 3 |
| chr19_+_43251539 | 0.00 |
ENSRNOT00000024793
|
Ddx19a
|
DEAD-box helicase 19A |
| chr20_-_4391402 | 0.00 |
ENSRNOT00000090518
ENSRNOT00000083489 |
Ppt2
|
palmitoyl-protein thioesterase 2 |
| chr3_+_125320680 | 0.00 |
ENSRNOT00000028891
|
LOC681292
|
hypothetical protein LOC681292 |
| chr3_-_113445435 | 0.00 |
ENSRNOT00000021562
|
Mfap1a
|
microfibrillar-associated protein 1A |
| chrM_+_10160 | 0.00 |
ENSRNOT00000042928
|
Mt-nd4
|
mitochondrially encoded NADH dehydrogenase 4 |
| chr9_-_94192813 | 0.00 |
ENSRNOT00000025978
|
Alpp
|
alkaline phosphatase, placental |
| chr8_-_117820413 | 0.00 |
ENSRNOT00000075819
|
Tma7
|
translation machinery associated 7 homolog |
| chr5_+_70528688 | 0.00 |
ENSRNOT00000036799
|
Fktn
|
fukutin |
| chr6_-_104409005 | 0.00 |
ENSRNOT00000040965
|
Ccdc177
|
coiled-coil domain containing 177 |
| chr1_-_228263198 | 0.00 |
ENSRNOT00000028572
|
Olr1874
|
olfactory receptor 1874 |
| chr10_-_76039964 | 0.00 |
ENSRNOT00000003164
|
Msi2
|
musashi RNA-binding protein 2 |
| chr14_+_3882398 | 0.00 |
ENSRNOT00000033794
|
Hfm1
|
HFM1, ATP-dependent DNA helicase homolog |
| chr1_-_47331412 | 0.00 |
ENSRNOT00000046746
|
Ezr
|
ezrin |
| chrM_-_14061 | 0.00 |
ENSRNOT00000051268
|
Mt-nd6
|
mitochondrially encoded NADH dehydrogenase 6 |
| chr13_+_94888078 | 0.00 |
ENSRNOT00000077995
ENSRNOT00000005647 ENSRNOT00000077345 |
Sdccag8
|
serologically defined colon cancer antigen 8 |
| chr5_-_150609352 | 0.00 |
ENSRNOT00000074374
|
Phactr4
|
phosphatase and actin regulator 4 |
| chr2_+_240642184 | 0.00 |
ENSRNOT00000083181
|
Ube2d3
|
ubiquitin-conjugating enzyme E2D 3 |
| chrX_-_17252877 | 0.00 |
ENSRNOT00000060213
|
LOC102547118
|
retinitis pigmentosa 1-like 1 protein-like |
| chr12_-_41497610 | 0.00 |
ENSRNOT00000001860
|
Iqcd
|
IQ motif containing D |
Network of associatons between targets according to the STRING database.
First level regulatory network of Otx1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:2000211 | regulation of glutamate metabolic process(GO:2000211) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:1903999 | negative regulation of eating behavior(GO:1903999) |
| 0.0 | 0.0 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.0 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.1 | GO:0033165 | interphotoreceptor matrix(GO:0033165) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.0 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |