Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
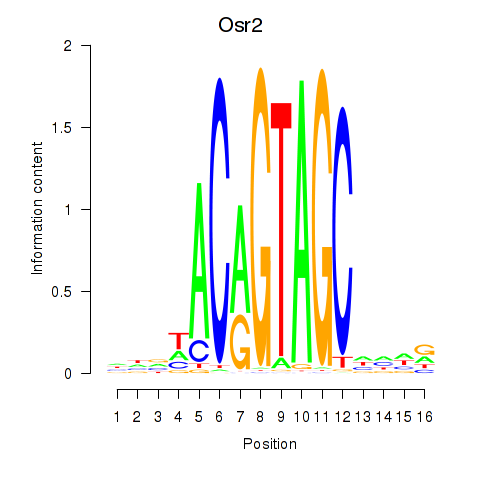
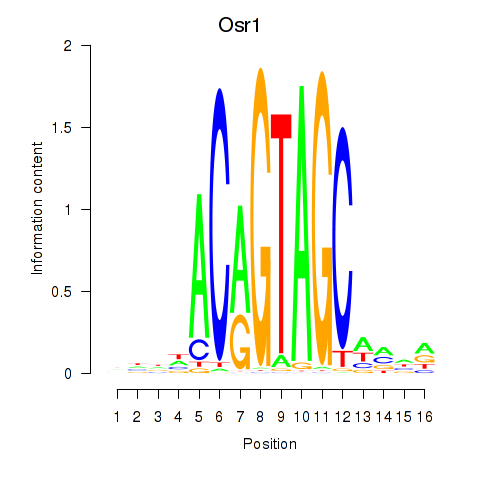
Results for Osr2_Osr1
Z-value: 0.55
Transcription factors associated with Osr2_Osr1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Osr2
|
ENSRNOG00000011136 | odd-skipped related transciption factor 2 |
|
Osr1
|
ENSRNOG00000004210 | odd-skipped related transciption factor 1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Osr1 | rn6_v1_chr6_+_35314444_35314444 | -0.72 | 1.7e-01 | Click! |
| Osr2 | rn6_v1_chr7_+_74047814_74047814 | -0.26 | 6.7e-01 | Click! |
Activity profile of Osr2_Osr1 motif
Sorted Z-values of Osr2_Osr1 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_48319522 | 0.25 |
ENSRNOT00000080281
|
AC114389.1
|
|
| chr4_-_181486090 | 0.25 |
ENSRNOT00000075453
|
AABR07062535.1
|
|
| chr8_+_48571323 | 0.23 |
ENSRNOT00000059776
|
Ccdc153
|
coiled-coil domain containing 153 |
| chr1_+_77782825 | 0.22 |
ENSRNOT00000058230
|
AABR07002623.1
|
|
| chr2_-_256154584 | 0.13 |
ENSRNOT00000072487
|
AABR07013776.1
|
|
| chr10_-_70788309 | 0.13 |
ENSRNOT00000029184
|
Ccl9
|
chemokine (C-C motif) ligand 9 |
| chr1_-_219438779 | 0.12 |
ENSRNOT00000029237
|
Tbc1d10c
|
TBC1 domain family, member 10C |
| chr11_+_67555658 | 0.10 |
ENSRNOT00000039075
|
Csta
|
cystatin A (stefin A) |
| chr14_-_114047527 | 0.10 |
ENSRNOT00000083199
|
AABR07016779.1
|
|
| chr1_-_164205222 | 0.10 |
ENSRNOT00000035774
|
Mogat2
|
monoacylglycerol O-acyltransferase 2 |
| chr3_-_151724654 | 0.10 |
ENSRNOT00000026964
|
Rbm39
|
RNA binding motif protein 39 |
| chr9_+_66889028 | 0.09 |
ENSRNOT00000087194
|
Carf
|
calcium responsive transcription factor |
| chr10_-_94500591 | 0.09 |
ENSRNOT00000015976
|
Cd79b
|
CD79b molecule |
| chr14_-_81339526 | 0.09 |
ENSRNOT00000015894
|
Grk4
|
G protein-coupled receptor kinase 4 |
| chr1_-_170578693 | 0.08 |
ENSRNOT00000025651
|
Rrp8
|
ribosomal RNA processing 8, methyltransferase, homolog (yeast) |
| chr8_-_55491152 | 0.08 |
ENSRNOT00000014965
|
LOC100359479
|
rCG58364-like |
| chr1_+_154131926 | 0.08 |
ENSRNOT00000035257
|
LOC102549852
|
ferritin light chain 1-like |
| chr5_-_2803855 | 0.08 |
ENSRNOT00000009490
|
LOC297756
|
ribosomal protein S8-like |
| chr5_+_74649765 | 0.08 |
ENSRNOT00000075952
|
Palm2
|
paralemmin 2 |
| chr19_-_39267928 | 0.07 |
ENSRNOT00000027686
|
Tmed6
|
transmembrane p24 trafficking protein 6 |
| chr2_-_35104963 | 0.07 |
ENSRNOT00000018058
|
Rgs7bp
|
regulator of G-protein signaling 7-binding protein |
| chr3_+_122816924 | 0.07 |
ENSRNOT00000045604
|
RGD1561317
|
similar to ribosomal protein L31 |
| chr2_+_58724855 | 0.07 |
ENSRNOT00000089609
|
Capsl
|
calcyphosine-like |
| chr16_-_69280109 | 0.07 |
ENSRNOT00000058595
|
AABR07026240.1
|
|
| chr6_+_127333590 | 0.07 |
ENSRNOT00000063970
|
Ifi27
|
interferon, alpha-inducible protein 27 |
| chr1_+_61522298 | 0.06 |
ENSRNOT00000029111
|
Zfp51
|
zinc finger protein 51 |
| chr1_-_167884690 | 0.06 |
ENSRNOT00000091372
|
Olr61
|
olfactory receptor 61 |
| chr18_+_62805410 | 0.06 |
ENSRNOT00000086679
|
Gnal
|
G protein subunit alpha L |
| chr6_+_137953545 | 0.06 |
ENSRNOT00000006804
|
Crip2
|
cysteine-rich protein 2 |
| chr7_+_144605058 | 0.06 |
ENSRNOT00000031404
|
Hoxc8
|
homeobox C8 |
| chr8_+_132869712 | 0.06 |
ENSRNOT00000008294
|
Cxcr6
|
C-X-C motif chemokine receptor 6 |
| chr11_-_31892531 | 0.05 |
ENSRNOT00000033745
|
Cryzl1
|
crystallin zeta like 1 |
| chr10_+_68173369 | 0.05 |
ENSRNOT00000037072
|
Tmem98
|
transmembrane protein 98 |
| chr2_+_147496229 | 0.05 |
ENSRNOT00000022105
|
Tm4sf4
|
transmembrane 4 L six family member 4 |
| chr5_-_163231494 | 0.05 |
ENSRNOT00000085929
ENSRNOT00000022968 |
Tnfrsf8
|
TNF receptor superfamily member 8 |
| chr5_+_154310453 | 0.05 |
ENSRNOT00000013322
|
Gale
|
UDP-galactose-4-epimerase |
| chr1_+_166564664 | 0.05 |
ENSRNOT00000090959
|
Pde2a
|
phosphodiesterase 2A |
| chr10_+_70153427 | 0.05 |
ENSRNOT00000076703
|
Lig3
|
DNA ligase 3 |
| chr11_+_38727048 | 0.05 |
ENSRNOT00000081537
|
LOC103690343
|
zinc finger protein 260-like |
| chr7_-_59139910 | 0.05 |
ENSRNOT00000032374
|
4933416C03Rik
|
RIKEN cDNA 4933416C03 gene |
| chrX_+_1543244 | 0.05 |
ENSRNOT00000071634
|
AABR07036746.1
|
|
| chr4_-_178441547 | 0.05 |
ENSRNOT00000087434
ENSRNOT00000055542 |
Sox5
|
SRY box 5 |
| chr14_-_6587141 | 0.05 |
ENSRNOT00000061664
|
LOC679894
|
similar to THO complex subunit 2 (Tho2) |
| chr3_+_119805941 | 0.05 |
ENSRNOT00000018584
|
Adra2b
|
adrenoceptor alpha 2B |
| chr6_+_144291974 | 0.05 |
ENSRNOT00000035255
|
Ncapg2
|
non-SMC condensin II complex, subunit G2 |
| chr7_-_145154131 | 0.05 |
ENSRNOT00000055271
|
Ppp1r1a
|
protein phosphatase 1, regulatory (inhibitor) subunit 1A |
| chr2_-_192671059 | 0.04 |
ENSRNOT00000012174
|
Sprr1a
|
small proline-rich protein 1A |
| chr5_-_17061361 | 0.04 |
ENSRNOT00000089318
|
Penk
|
proenkephalin |
| chr10_+_86223336 | 0.04 |
ENSRNOT00000038939
|
LOC257650
|
hippyragranin |
| chr17_-_14687408 | 0.04 |
ENSRNOT00000088542
|
AC120310.1
|
|
| chr15_-_40545824 | 0.04 |
ENSRNOT00000017204
|
Nup58
|
nucleoporin 58 |
| chr10_+_95242986 | 0.04 |
ENSRNOT00000065589
ENSRNOT00000080460 |
RGD1565033
|
similar to hypothetical protein LOC284018 isoform b |
| chr1_-_143169657 | 0.04 |
ENSRNOT00000025888
|
Rps17
|
ribosomal protein S17 |
| chr6_-_33088582 | 0.04 |
ENSRNOT00000083144
|
Tdrd15
|
tudor domain containing 15 |
| chrX_+_123350346 | 0.04 |
ENSRNOT00000017026
|
Slc25a43
|
solute carrier family 25, member 43 |
| chr3_+_138597638 | 0.04 |
ENSRNOT00000031623
|
Zfp133
|
zinc finger protein 133 |
| chr1_-_250626844 | 0.04 |
ENSRNOT00000077135
|
Asah2
|
N-acylsphingosine amidohydrolase 2 |
| chr20_-_31736212 | 0.04 |
ENSRNOT00000000662
|
RGD1305587
|
similar to RIKEN cDNA 2010107G23 |
| chr15_+_105640097 | 0.04 |
ENSRNOT00000014300
|
Mbnl2
|
muscleblind-like splicing regulator 2 |
| chr15_-_60289763 | 0.04 |
ENSRNOT00000038579
|
Fam216b
|
family with sequence similarity 216, member B |
| chr8_+_50200069 | 0.04 |
ENSRNOT00000023677
|
Pcsk7
|
proprotein convertase subtilisin/kexin type 7 |
| chr8_+_107875991 | 0.04 |
ENSRNOT00000090299
|
Dzip1l
|
DAZ interacting zinc finger protein 1-like |
| chr2_+_4942775 | 0.04 |
ENSRNOT00000093548
ENSRNOT00000093741 |
Fam172a
|
family with sequence similarity 172, member A |
| chrM_+_8599 | 0.03 |
ENSRNOT00000049683
|
Mt-cox3
|
mitochondrially encoded cytochrome C oxidase III |
| chr7_+_26375866 | 0.03 |
ENSRNOT00000059639
|
Aldh1l2
|
aldehyde dehydrogenase 1 family, member L2 |
| chr9_+_67774150 | 0.03 |
ENSRNOT00000091060
|
Icos
|
inducible T-cell co-stimulator |
| chr2_-_45077219 | 0.03 |
ENSRNOT00000014319
|
Gzmk
|
granzyme K |
| chr5_+_18901039 | 0.03 |
ENSRNOT00000012066
|
Fam110b
|
family with sequence similarity 110, member B |
| chr2_+_187218851 | 0.03 |
ENSRNOT00000017798
|
Sh2d2a
|
SH2 domain containing 2A |
| chr2_-_26011429 | 0.03 |
ENSRNOT00000065143
|
Arhgef28
|
Rho guanine nucleotide exchange factor 28 |
| chr10_-_56276764 | 0.03 |
ENSRNOT00000049048
|
Eif4a1
|
eukaryotic translation initiation factor 4A1 |
| chr1_-_80483487 | 0.03 |
ENSRNOT00000074418
|
Gemin7
|
gem (nuclear organelle) associated protein 7 |
| chr10_+_3411380 | 0.03 |
ENSRNOT00000004346
|
RGD1305733
|
similar to RIKEN cDNA 2900011O08 |
| chr14_+_19319299 | 0.03 |
ENSRNOT00000086542
|
Ankrd17
|
ankyrin repeat domain 17 |
| chr1_+_99706780 | 0.03 |
ENSRNOT00000024758
|
Klk12
|
kallikrein related-peptidase 12 |
| chr14_+_71533460 | 0.03 |
ENSRNOT00000084274
|
Prom1
|
prominin 1 |
| chr8_-_64800467 | 0.03 |
ENSRNOT00000073122
|
Nr2e3
|
nuclear receptor subfamily 2, group E, member 3 |
| chr17_+_69634890 | 0.03 |
ENSRNOT00000029049
|
Akr1c13
|
aldo-keto reductase family 1, member C13 |
| chr1_-_167893061 | 0.03 |
ENSRNOT00000049401
|
Olr60
|
olfactory receptor 60 |
| chr16_-_20864176 | 0.03 |
ENSRNOT00000092706
|
Cope
|
coatomer protein complex, subunit epsilon |
| chr20_-_11528332 | 0.03 |
ENSRNOT00000038419
|
Tspear
|
thrombospondin-type laminin G domain and EAR repeats |
| chr9_-_99818262 | 0.03 |
ENSRNOT00000056600
|
Cops9
|
COP9 signalosome subunit 9 |
| chr4_-_121565212 | 0.03 |
ENSRNOT00000088753
|
Chchd6
|
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr4_+_57855416 | 0.03 |
ENSRNOT00000029608
|
Cpa2
|
carboxypeptidase A2 |
| chr3_+_113976687 | 0.03 |
ENSRNOT00000022437
|
Eif3j
|
eukaryotic translation initiation factor 3, subunit J |
| chr8_-_36467627 | 0.03 |
ENSRNOT00000082346
|
Fam118b
|
family with sequence similarity 118, member B |
| chr4_-_181348038 | 0.03 |
ENSRNOT00000082879
|
LOC690784
|
hypothetical protein LOC690783 |
| chr10_+_7041510 | 0.03 |
ENSRNOT00000003514
|
Carhsp1
|
calcium regulated heat stable protein 1 |
| chr8_+_72029489 | 0.03 |
ENSRNOT00000089336
|
Herc1
|
HECT and RLD domain containing E3 ubiquitin protein ligase family member 1 |
| chr18_-_58423196 | 0.03 |
ENSRNOT00000025556
|
Piezo2
|
piezo-type mechanosensitive ion channel component 2 |
| chr15_-_100348760 | 0.03 |
ENSRNOT00000085607
|
AABR07019341.1
|
|
| chr12_-_30491102 | 0.03 |
ENSRNOT00000001224
|
Sumf2
|
sulfatase modifying factor 2 |
| chr11_+_82862695 | 0.03 |
ENSRNOT00000071707
ENSRNOT00000073435 |
Clcn2
|
chloride voltage-gated channel 2 |
| chr11_-_27080701 | 0.03 |
ENSRNOT00000002180
|
Ltn1
|
listerin E3 ubiquitin protein ligase 1 |
| chr11_+_83883879 | 0.03 |
ENSRNOT00000050927
|
LOC108348101
|
chloride channel protein 2 |
| chr7_-_102298522 | 0.03 |
ENSRNOT00000006273
|
A1bg
|
alpha-1-B glycoprotein |
| chrX_+_53053609 | 0.03 |
ENSRNOT00000058357
|
Dmd
|
dystrophin |
| chr5_+_70592871 | 0.02 |
ENSRNOT00000035687
|
Tal2
|
TAL bHLH transcription factor 2 |
| chr10_-_109278510 | 0.02 |
ENSRNOT00000038993
|
Tepsin
|
TEPSIN, adaptor related protein complex 4 accessory protein |
| chr1_-_212568224 | 0.02 |
ENSRNOT00000024976
|
LOC100911225
|
fucose mutarotase-like |
| chr18_-_48384645 | 0.02 |
ENSRNOT00000023485
|
Ppic
|
peptidylprolyl isomerase C |
| chr1_-_53087474 | 0.02 |
ENSRNOT00000017302
|
Ccr6
|
C-C motif chemokine receptor 6 |
| chr1_+_163398891 | 0.02 |
ENSRNOT00000020513
|
Gucy2e
|
guanylate cyclase 2E |
| chr4_+_68849033 | 0.02 |
ENSRNOT00000016912
|
Mgam
|
maltase-glucoamylase |
| chr16_+_10277775 | 0.02 |
ENSRNOT00000090679
|
Rbp3
|
retinol binding protein 3 |
| chr5_+_8876044 | 0.02 |
ENSRNOT00000008877
|
Cops5
|
COP9 signalosome subunit 5 |
| chr1_-_163148948 | 0.02 |
ENSRNOT00000046850
|
B3gnt6
|
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 6 |
| chr9_+_98113346 | 0.02 |
ENSRNOT00000026912
|
Prlh
|
prolactin releasing hormone |
| chr1_-_260460791 | 0.02 |
ENSRNOT00000018179
|
Tll2
|
tolloid-like 2 |
| chrX_-_15761932 | 0.02 |
ENSRNOT00000015641
ENSRNOT00000091146 |
Foxp3
|
forkhead box P3 |
| chr1_+_101610773 | 0.02 |
ENSRNOT00000032535
|
Rasip1
|
Ras interacting protein 1 |
| chr8_+_115131367 | 0.02 |
ENSRNOT00000014849
|
Rpl29
|
ribosomal protein L29 |
| chr14_+_84482674 | 0.02 |
ENSRNOT00000009313
|
Lif
|
leukemia inhibitory factor |
| chr10_-_15282951 | 0.02 |
ENSRNOT00000027266
|
Rab40c
|
Rab40c, member RAS oncogene family |
| chr14_+_2050483 | 0.02 |
ENSRNOT00000000047
|
Slc26a1
|
solute carrier family 26 member 1 |
| chr2_+_247299433 | 0.02 |
ENSRNOT00000050330
|
Unc5c
|
unc-5 netrin receptor C |
| chr1_+_260153645 | 0.02 |
ENSRNOT00000054717
|
Zfp518a
|
zinc finger protein 518A |
| chr17_+_44522140 | 0.02 |
ENSRNOT00000080490
|
RGD1562378
|
histone H4 variant H4-v.1 |
| chr4_-_163157744 | 0.02 |
ENSRNOT00000089712
|
Clec12b
|
C-type lectin domain family 12, member B |
| chr15_-_18675431 | 0.02 |
ENSRNOT00000080794
|
Abhd6
|
abhydrolase domain containing 6 |
| chr7_+_64987859 | 0.02 |
ENSRNOT00000005694
|
Tmbim4
|
transmembrane BAX inhibitor motif containing 4 |
| chr4_-_133127175 | 0.02 |
ENSRNOT00000057758
|
Shq1
|
SHQ1, H/ACA ribonucleoprotein assembly factor |
| chr11_-_1819094 | 0.02 |
ENSRNOT00000000911
|
MGC95208
|
similar to 4930453N24Rik protein |
| chr3_+_170252901 | 0.02 |
ENSRNOT00000005871
|
Mc3r
|
melanocortin 3 receptor |
| chr7_-_36408588 | 0.02 |
ENSRNOT00000063946
|
Cradd
|
CASP2 and RIPK1 domain containing adaptor with death domain |
| chr6_+_137386422 | 0.02 |
ENSRNOT00000018568
|
RGD1307315
|
LOC362793 |
| chr11_+_83884048 | 0.02 |
ENSRNOT00000002376
|
LOC108348101
|
chloride channel protein 2 |
| chr4_-_50200328 | 0.02 |
ENSRNOT00000060530
|
Aass
|
aminoadipate-semialdehyde synthase |
| chr1_+_141832774 | 0.02 |
ENSRNOT00000073302
|
Fuom
|
fucose mutarotase |
| chr12_+_42097626 | 0.02 |
ENSRNOT00000001893
|
Tbx5
|
T-box 5 |
| chr8_-_112807598 | 0.02 |
ENSRNOT00000015344
|
Dnajc13
|
DnaJ heat shock protein family (Hsp40) member C13 |
| chr3_-_152259156 | 0.02 |
ENSRNOT00000065729
|
LOC100910882
|
RNA-binding protein 39-like |
| chr9_-_11080155 | 0.02 |
ENSRNOT00000072830
|
Fsd1
|
fibronectin type III and SPRY domain containing 1 |
| chr10_-_56850085 | 0.02 |
ENSRNOT00000025767
|
Rnasek
|
ribonuclease K |
| chr19_-_53477420 | 0.02 |
ENSRNOT00000071344
|
LOC687560
|
hypothetical protein LOC687560 |
| chr10_+_15697227 | 0.02 |
ENSRNOT00000028008
|
Il9r
|
interleukin 9 receptor |
| chr1_-_240601744 | 0.02 |
ENSRNOT00000024093
|
Aldh1a7
|
aldehyde dehydrogenase family 1, subfamily A7 |
| chr3_-_150412179 | 0.02 |
ENSRNOT00000023786
|
Eif2s2
|
eukaryotic translation initiation factor 2 subunit beta |
| chr12_-_19439977 | 0.02 |
ENSRNOT00000060035
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr15_+_18451144 | 0.01 |
ENSRNOT00000010260
|
Acox2
|
acyl-CoA oxidase 2 |
| chr6_-_11274932 | 0.01 |
ENSRNOT00000021538
|
Msh2
|
mutS homolog 2 |
| chr18_+_59748444 | 0.01 |
ENSRNOT00000024752
|
St8sia3
|
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 |
| chr16_-_76110553 | 0.01 |
ENSRNOT00000043384
|
Mcph1
|
microcephalin 1 |
| chr2_+_196655469 | 0.01 |
ENSRNOT00000028730
|
Ctsk
|
cathepsin K |
| chr15_-_51400606 | 0.01 |
ENSRNOT00000023318
|
Chmp7
|
charged multivesicular body protein 7 |
| chr12_+_19513100 | 0.01 |
ENSRNOT00000030161
|
RGD1562319
|
similar to family with sequence similarity 55, member C |
| chr12_-_19440501 | 0.01 |
ENSRNOT00000060048
|
Nxpe5
|
neurexophilin and PC-esterase domain family, member 5 |
| chr12_+_52397792 | 0.01 |
ENSRNOT00000085661
ENSRNOT00000056694 ENSRNOT00000056693 ENSRNOT00000056692 ENSRNOT00000081447 |
P2rx2
|
purinergic receptor P2X 2 |
| chr4_+_38255413 | 0.01 |
ENSRNOT00000089897
|
Phf14
|
PHD finger protein 14 |
| chr4_-_121684264 | 0.01 |
ENSRNOT00000083168
|
Vom1r90
|
vomeronasal 1 receptor 90 |
| chr3_-_13865843 | 0.01 |
ENSRNOT00000025194
|
Rabepk
|
Rab9 effector protein with kelch motifs |
| chr9_+_90857308 | 0.01 |
ENSRNOT00000073993
|
LOC100911572
|
collagen alpha-3(IV) chain-like |
| chrX_+_156355376 | 0.01 |
ENSRNOT00000078304
|
Lage3
|
L antigen family, member 3 |
| chr10_-_59049482 | 0.01 |
ENSRNOT00000078272
|
Spns2
|
spinster homolog 2 |
| chr10_-_16731898 | 0.01 |
ENSRNOT00000028186
|
Crebrf
|
CREB3 regulatory factor |
| chr13_+_106751625 | 0.01 |
ENSRNOT00000004992
|
Ush2a
|
usherin |
| chr1_+_91363492 | 0.01 |
ENSRNOT00000014517
|
Cebpa
|
CCAAT/enhancer binding protein alpha |
| chr7_-_57816621 | 0.01 |
ENSRNOT00000090748
|
AABR07057150.1
|
|
| chr1_+_170578889 | 0.01 |
ENSRNOT00000025906
|
Ilk
|
integrin-linked kinase |
| chr20_-_32076503 | 0.01 |
ENSRNOT00000000441
|
Supv3l1
|
Suv3 like RNA helicase |
| chr1_-_78573374 | 0.01 |
ENSRNOT00000090519
|
Arhgap35
|
Rho GTPase activating protein 35 |
| chr3_+_72395218 | 0.01 |
ENSRNOT00000057616
|
Prg3
|
proteoglycan 3, pro eosinophil major basic protein 2 |
| chr5_+_64053946 | 0.01 |
ENSRNOT00000011622
|
Invs
|
inversin |
| chrX_+_68626185 | 0.01 |
ENSRNOT00000090942
|
Yipf6
|
Yip1 domain family, member 6 |
| chr8_-_45137893 | 0.01 |
ENSRNOT00000010743
|
RGD1309108
|
similar to hypothetical protein FLJ23554 |
| chr1_-_128604188 | 0.01 |
ENSRNOT00000033858
|
Lrrc28
|
leucine rich repeat containing 28 |
| chr6_+_8284878 | 0.01 |
ENSRNOT00000009581
|
Slc3a1
|
solute carrier family 3 member 1 |
| chr4_-_62663200 | 0.01 |
ENSRNOT00000040969
|
Cnot4
|
CCR4-NOT transcription complex, subunit 4 |
| chr10_-_56849255 | 0.01 |
ENSRNOT00000025491
|
RGD1308134
|
similar to RIKEN cDNA 1110020A23 |
| chr1_-_91588609 | 0.01 |
ENSRNOT00000050931
|
Snrpg
|
small nuclear ribonucleoprotein polypeptide G |
| chr2_-_149325913 | 0.01 |
ENSRNOT00000036690
|
Gpr171
|
G protein-coupled receptor 171 |
| chrX_-_77956352 | 0.01 |
ENSRNOT00000003296
|
Zcchc5
|
zinc finger CCHC-type containing 5 |
| chr11_+_34316295 | 0.01 |
ENSRNOT00000081834
|
Sim2
|
single-minded family bHLH transcription factor 2 |
| chr2_+_237679792 | 0.01 |
ENSRNOT00000064628
|
Gimd1
|
GIMAP family P-loop NTPase domain containing 1 |
| chr2_-_5577369 | 0.01 |
ENSRNOT00000093420
|
Nr2f1
|
nuclear receptor subfamily 2, group F, member 1 |
| chr13_+_101698096 | 0.01 |
ENSRNOT00000089309
|
Aida
|
axin interactor, dorsalization associated |
| chr1_-_73682247 | 0.01 |
ENSRNOT00000079498
|
Lilra5
|
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 5 |
| chr19_-_29802083 | 0.01 |
ENSRNOT00000024755
|
AC123471.1
|
|
| chr1_+_70222964 | 0.01 |
ENSRNOT00000050180
|
Peg3
|
paternally expressed 3 |
| chr15_+_344685 | 0.01 |
ENSRNOT00000065542
ENSRNOT00000066928 |
Kcnma1
|
potassium calcium-activated channel subfamily M alpha 1 |
| chr9_-_11027506 | 0.01 |
ENSRNOT00000071107
|
Chaf1a
|
chromatin assembly factor 1 subunit A |
| chr1_+_218532045 | 0.01 |
ENSRNOT00000018848
|
Mrpl21
|
mitochondrial ribosomal protein L21 |
| chr3_+_72134731 | 0.01 |
ENSRNOT00000083592
|
Ypel4
|
yippee-like 4 |
| chr7_+_37101391 | 0.01 |
ENSRNOT00000029764
|
Eea1
|
early endosome antigen 1 |
| chr2_-_80408672 | 0.01 |
ENSRNOT00000067638
|
Fam105a
|
family with sequence similarity 105, member A |
| chr10_+_34383396 | 0.01 |
ENSRNOT00000047186
|
Olr1386
|
olfactory receptor 1386 |
| chr10_+_11240138 | 0.01 |
ENSRNOT00000048687
|
Srl
|
sarcalumenin |
| chr2_-_188736462 | 0.01 |
ENSRNOT00000028030
|
Flad1
|
flavin adenine dinucleotide synthetase 1 |
| chrX_-_83151511 | 0.01 |
ENSRNOT00000057378
|
Hdx
|
highly divergent homeobox |
| chr17_-_10061750 | 0.01 |
ENSRNOT00000082042
ENSRNOT00000022646 |
Zfp346
|
zinc finger protein 346 |
| chr11_+_71222600 | 0.01 |
ENSRNOT00000002418
|
Muc20
|
mucin 20, cell surface associated |
| chr16_+_53998560 | 0.01 |
ENSRNOT00000077188
ENSRNOT00000013463 |
Asah1
|
N-acylsphingosine amidohydrolase 1 |
| chrX_-_75566481 | 0.01 |
ENSRNOT00000003714
|
Zdhhc15
|
zinc finger, DHHC-type containing 15 |
| chr2_+_45104305 | 0.01 |
ENSRNOT00000014559
|
Esm1
|
endothelial cell-specific molecule 1 |
| chr8_-_50199978 | 0.01 |
ENSRNOT00000023459
|
Rnf214
|
ring finger protein 214 |
| chr6_+_128738388 | 0.01 |
ENSRNOT00000050204
|
Rpl6-ps1
|
ribosomal protein L6, pseudogene 1 |
| chr18_-_27395603 | 0.01 |
ENSRNOT00000034385
|
LOC100359679
|
rCG49431-like |
| chr12_-_52396066 | 0.01 |
ENSRNOT00000056712
|
Lrcol1
|
leucine rich colipase-like 1 |
| chr1_-_142183884 | 0.01 |
ENSRNOT00000016032
|
Fes
|
FES proto-oncogene, tyrosine kinase |
| chr2_-_139528162 | 0.01 |
ENSRNOT00000014317
|
Slc7a11
|
solute carrier family 7 member 11 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Osr2_Osr1
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0032759 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.0 | GO:0006288 | base-excision repair, DNA ligation(GO:0006288) |
| 0.0 | 0.0 | GO:0051867 | general adaptation syndrome, behavioral process(GO:0051867) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.0 | GO:0035625 | receptor transactivation(GO:0035624) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) activation of MAPK activity by adrenergic receptor signaling pathway(GO:0071883) |
| 0.0 | 0.1 | GO:0046015 | regulation of transcription by glucose(GO:0046015) |
| 0.0 | 0.0 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0044302 | dentate gyrus mossy fiber(GO:0044302) |
| 0.0 | 0.0 | GO:0070421 | DNA ligase III-XRCC1 complex(GO:0070421) |
| 0.0 | 0.1 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.1 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0003846 | 2-acylglycerol O-acyltransferase activity(GO:0003846) |
| 0.0 | 0.0 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.0 | 0.1 | GO:0036004 | GAF domain binding(GO:0036004) |
| 0.0 | 0.1 | GO:0004703 | G-protein coupled receptor kinase activity(GO:0004703) |
Gene overrepresentation in curated gene sets: REACTOME pathways category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |