Project
GSE49485: Hypoxia transcriptome sequencing of rat brain.
Navigation
Downloads
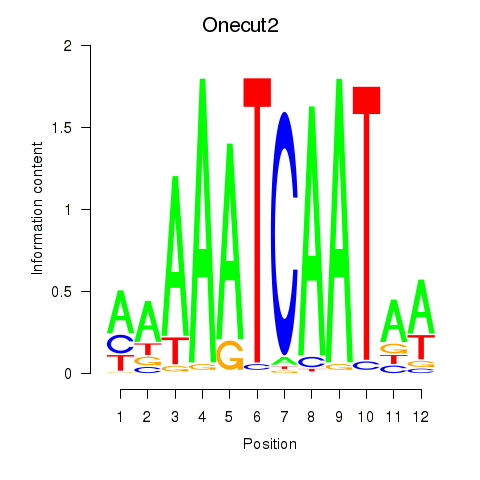
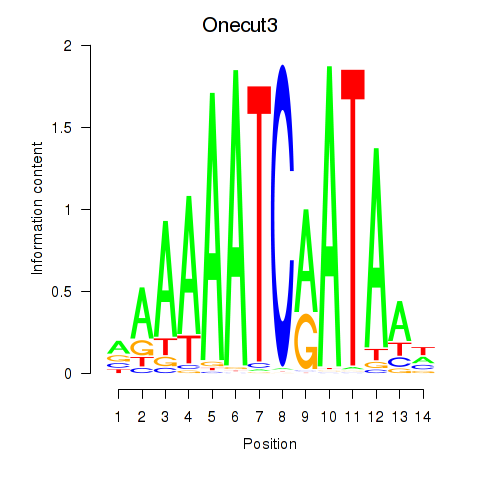
Results for Onecut2_Onecut3
Z-value: 0.23
Transcription factors associated with Onecut2_Onecut3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
Onecut2
|
ENSRNOG00000052665 | one cut domain, family member 2 |
|
Onecut3
|
ENSRNOG00000059889 | one cut domain, family member 3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| Onecut3 | rn6_v1_chr7_-_12085226_12085226 | 0.71 | 1.8e-01 | Click! |
| Onecut2 | rn6_v1_chr18_+_59830363_59830422 | -0.36 | 5.5e-01 | Click! |
Activity profile of Onecut2_Onecut3 motif
Sorted Z-values of Onecut2_Onecut3 motif
| Promoter | Log-likelihood | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_190370499 | 0.26 |
ENSRNOT00000084389
|
AABR07005618.1
|
|
| chr10_+_61685645 | 0.15 |
ENSRNOT00000003933
|
Mnt
|
MAX network transcriptional repressor |
| chr2_+_27905535 | 0.13 |
ENSRNOT00000022120
|
Fam169a
|
family with sequence similarity 169, member A |
| chr5_-_153840178 | 0.11 |
ENSRNOT00000025075
|
Nipal3
|
NIPA-like domain containing 3 |
| chr5_-_168734296 | 0.10 |
ENSRNOT00000066120
|
Camta1
|
calmodulin binding transcription activator 1 |
| chr5_-_127878792 | 0.09 |
ENSRNOT00000014436
|
Zyg11b
|
zyg-11 family member B, cell cycle regulator |
| chr7_+_34533543 | 0.09 |
ENSRNOT00000007412
ENSRNOT00000084185 |
Ntn4
|
netrin 4 |
| chr10_+_35343189 | 0.08 |
ENSRNOT00000083688
|
Mapk9
|
mitogen-activated protein kinase 9 |
| chr18_-_77579969 | 0.08 |
ENSRNOT00000034896
|
Sall3
|
spalt-like transcription factor 3 |
| chr1_-_124803363 | 0.08 |
ENSRNOT00000066380
|
Klf13
|
Kruppel-like factor 13 |
| chr16_+_85331866 | 0.07 |
ENSRNOT00000019615
|
Lig4
|
DNA ligase 4 |
| chr7_-_114848414 | 0.07 |
ENSRNOT00000032620
|
Slc45a4
|
solute carrier family 45, member 4 |
| chr5_+_76092287 | 0.06 |
ENSRNOT00000020207
|
Zfp483
|
zinc finger protein 483 |
| chr13_-_73055631 | 0.06 |
ENSRNOT00000081892
|
Xpr1
|
xenotropic and polytropic retrovirus receptor 1 |
| chr2_-_39007976 | 0.06 |
ENSRNOT00000064292
|
Zswim6
|
zinc finger, SWIM-type containing 6 |
| chr9_-_17880706 | 0.06 |
ENSRNOT00000031549
|
Aars2
|
alanyl-tRNA synthetase 2, mitochondrial |
| chr7_+_3216497 | 0.06 |
ENSRNOT00000008909
|
Mmp19
|
matrix metallopeptidase 19 |
| chr13_+_82231030 | 0.06 |
ENSRNOT00000003551
|
Scyl3
|
SCY1 like pseudokinase 3 |
| chr5_-_105579959 | 0.06 |
ENSRNOT00000010827
|
Slc24a2
|
solute carrier family 24 member 2 |
| chr1_-_175657485 | 0.05 |
ENSRNOT00000024561
|
rnf141
|
ring finger protein 141 |
| chr10_+_32824 | 0.05 |
ENSRNOT00000077828
|
LOC103689943
|
meiosis arrest female protein 1-like |
| chr2_-_231409496 | 0.05 |
ENSRNOT00000055615
ENSRNOT00000015386 ENSRNOT00000068415 |
Ank2
|
ankyrin 2 |
| chr8_+_76754492 | 0.05 |
ENSRNOT00000085727
|
Myo1e
|
myosin IE |
| chr7_+_42304534 | 0.05 |
ENSRNOT00000085097
|
Kitlg
|
KIT ligand |
| chrX_+_120901495 | 0.05 |
ENSRNOT00000040742
|
Wdr44
|
WD repeat domain 44 |
| chr10_-_91986632 | 0.05 |
ENSRNOT00000087824
|
Nsf
|
N-ethylmaleimide sensitive factor, vesicle fusing ATPase |
| chr10_-_27179254 | 0.05 |
ENSRNOT00000004619
|
Gabrg2
|
gamma-aminobutyric acid type A receptor gamma 2 subunit |
| chr14_-_84751886 | 0.04 |
ENSRNOT00000078838
|
Mtmr3
|
myotubularin related protein 3 |
| chr6_-_41870046 | 0.04 |
ENSRNOT00000005863
|
Lpin1
|
lipin 1 |
| chr14_-_16903242 | 0.04 |
ENSRNOT00000003001
|
Shroom3
|
shroom family member 3 |
| chr3_+_79823945 | 0.04 |
ENSRNOT00000014484
|
Celf1
|
CUGBP, Elav-like family member 1 |
| chr12_-_45801842 | 0.04 |
ENSRNOT00000078837
|
AABR07036513.1
|
|
| chr1_-_197568165 | 0.04 |
ENSRNOT00000079044
|
Xpo6
|
exportin 6 |
| chr10_-_15465404 | 0.04 |
ENSRNOT00000077826
ENSRNOT00000027593 |
Decr2
|
2,4-dienoyl-CoA reductase 2 |
| chr3_+_8928534 | 0.04 |
ENSRNOT00000088509
ENSRNOT00000065300 |
Miga2
|
mitoguardin 2 |
| chr18_+_30496318 | 0.04 |
ENSRNOT00000027179
|
Pcdhb11
|
protocadherin beta 11 |
| chr8_-_22821397 | 0.04 |
ENSRNOT00000045488
|
Kank2
|
KN motif and ankyrin repeat domains 2 |
| chr8_-_63750531 | 0.04 |
ENSRNOT00000009496
|
Neo1
|
neogenin 1 |
| chr2_+_186980793 | 0.04 |
ENSRNOT00000091336
|
Arhgef11
|
Rho guanine nucleotide exchange factor 11 |
| chr2_+_87018305 | 0.04 |
ENSRNOT00000079449
|
Rsl1
|
regulator of sex limited protein 1 |
| chr17_-_4454701 | 0.04 |
ENSRNOT00000080750
ENSRNOT00000066950 |
Dapk1
|
death associated protein kinase 1 |
| chr19_+_39543242 | 0.04 |
ENSRNOT00000018837
|
Wwp2
|
WW domain containing E3 ubiquitin protein ligase 2 |
| chr1_+_98627372 | 0.04 |
ENSRNOT00000030370
|
Dzf17
|
zinc finger protein 17 |
| chr5_+_187312 | 0.04 |
ENSRNOT00000030857
|
LOC102557117
|
zinc finger protein 120-like |
| chr9_+_6970507 | 0.04 |
ENSRNOT00000079488
|
St6gal2
|
ST6 beta-galactoside alpha-2,6-sialyltransferase 2 |
| chr13_+_90260783 | 0.03 |
ENSRNOT00000050547
|
Cd84
|
CD84 molecule |
| chr1_+_84685931 | 0.03 |
ENSRNOT00000063839
|
Zfp780b
|
zinc finger protein 780B |
| chr2_-_196582997 | 0.03 |
ENSRNOT00000081672
|
AABR07012475.1
|
|
| chr2_+_113007549 | 0.03 |
ENSRNOT00000017758
|
Tnfsf10
|
tumor necrosis factor superfamily member 10 |
| chr3_+_11653529 | 0.03 |
ENSRNOT00000091048
|
Ak1
|
adenylate kinase 1 |
| chr20_-_31597830 | 0.03 |
ENSRNOT00000085877
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr3_+_67870111 | 0.03 |
ENSRNOT00000012628
|
Nup35
|
nucleoporin 35 |
| chr2_+_200603426 | 0.03 |
ENSRNOT00000072189
|
LOC102550892
|
zinc finger protein 697-like |
| chr3_+_116899878 | 0.03 |
ENSRNOT00000090802
ENSRNOT00000066101 |
Sema6d
|
semaphorin 6D |
| chr14_-_104960141 | 0.03 |
ENSRNOT00000056944
|
Aftph
|
aftiphilin |
| chr18_-_37776453 | 0.03 |
ENSRNOT00000087876
|
Dpysl3
|
dihydropyrimidinase-like 3 |
| chr14_-_82171480 | 0.03 |
ENSRNOT00000021952
|
Nsd2
|
nuclear receptor binding SET domain protein 2 |
| chr15_-_14737704 | 0.03 |
ENSRNOT00000011307
|
Synpr
|
synaptoporin |
| chr2_+_252771017 | 0.03 |
ENSRNOT00000075452
ENSRNOT00000055318 |
Ttll7
|
tubulin tyrosine ligase like 7 |
| chr1_-_255815733 | 0.03 |
ENSRNOT00000047387
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3 |
| chr2_+_86891092 | 0.03 |
ENSRNOT00000077162
ENSRNOT00000083066 |
LOC100360380
|
zinc finger protein 457-like |
| chr2_+_195996521 | 0.03 |
ENSRNOT00000082849
|
Pogz
|
pogo transposable element with ZNF domain |
| chr2_+_104020955 | 0.03 |
ENSRNOT00000045586
|
Mtfr1
|
mitochondrial fission regulator 1 |
| chr20_-_31598118 | 0.03 |
ENSRNOT00000046537
|
Col13a1
|
collagen type XIII alpha 1 chain |
| chr7_-_81592206 | 0.03 |
ENSRNOT00000007979
|
Angpt1
|
angiopoietin 1 |
| chr1_-_263885169 | 0.03 |
ENSRNOT00000030782
|
Chuk
|
conserved helix-loop-helix ubiquitous kinase |
| chr3_-_83306781 | 0.03 |
ENSRNOT00000014088
|
Ttc17
|
tetratricopeptide repeat domain 17 |
| chr2_-_57600820 | 0.03 |
ENSRNOT00000083247
|
Nipbl
|
NIPBL, cohesin loading factor |
| chr3_-_158328881 | 0.03 |
ENSRNOT00000044466
|
Ptprt
|
protein tyrosine phosphatase, receptor type, T |
| chrX_-_84821775 | 0.02 |
ENSRNOT00000000174
|
Chm
|
CHM, Rab escort protein 1 |
| chr3_+_129753742 | 0.02 |
ENSRNOT00000007998
ENSRNOT00000080581 |
Snap25
|
synaptosomal-associated protein 25 |
| chr5_-_77031120 | 0.02 |
ENSRNOT00000022987
|
Inip
|
INTS3 and NABP interacting protein |
| chr4_+_64088900 | 0.02 |
ENSRNOT00000075341
|
Chrm2
|
cholinergic receptor, muscarinic 2 |
| chr9_-_15274917 | 0.02 |
ENSRNOT00000019650
|
Pgc
|
progastricsin |
| chr14_-_103321270 | 0.02 |
ENSRNOT00000006157
|
Meis1
|
Meis homeobox 1 |
| chr10_-_71358190 | 0.02 |
ENSRNOT00000078646
|
Ac1576
|
uncharacterized LOC102552783 |
| chrX_+_65566047 | 0.02 |
ENSRNOT00000092103
|
Heph
|
hephaestin |
| chr19_-_38484611 | 0.02 |
ENSRNOT00000077960
|
Nfat5
|
nuclear factor of activated T-cells 5 |
| chr10_+_80953006 | 0.02 |
ENSRNOT00000035567
|
Hnrnpa3
|
heterogeneous nuclear ribonucleoprotein A3 |
| chrX_+_6273733 | 0.02 |
ENSRNOT00000074275
|
Ndp
|
NDP, norrin cystine knot growth factor |
| chr2_+_235596907 | 0.02 |
ENSRNOT00000071463
ENSRNOT00000075728 |
Col25a1
|
collagen type XXV alpha 1 chain |
| chr10_+_10808823 | 0.02 |
ENSRNOT00000004298
|
Anks3
|
ankyrin repeat and sterile alpha motif domain containing 3 |
| chr2_+_116032455 | 0.02 |
ENSRNOT00000066495
|
Phc3
|
polyhomeotic homolog 3 |
| chr15_-_52288893 | 0.02 |
ENSRNOT00000016209
|
Fam160b2
|
family with sequence similarity 160, member B2 |
| chr17_+_8135251 | 0.02 |
ENSRNOT00000016982
|
Trpc7
|
transient receptor potential cation channel, subfamily C, member 7 |
| chr16_-_10603850 | 0.02 |
ENSRNOT00000087737
|
Fam35a
|
family with sequence similarity 35, member A |
| chr2_+_27000327 | 0.02 |
ENSRNOT00000087418
|
Poc5
|
POC5 centriolar protein |
| chr10_+_65767053 | 0.02 |
ENSRNOT00000078897
|
Vtn
|
vitronectin |
| chr1_+_80028928 | 0.02 |
ENSRNOT00000012082
|
Fbxo46
|
F-box protein 46 |
| chr15_-_28746042 | 0.02 |
ENSRNOT00000017730
|
Sall2
|
spalt-like transcription factor 2 |
| chr8_-_80631873 | 0.02 |
ENSRNOT00000091661
ENSRNOT00000080662 |
Unc13c
|
unc-13 homolog C |
| chr2_-_38110567 | 0.02 |
ENSRNOT00000072212
|
Ipo11
|
importin 11 |
| chr8_+_70317586 | 0.02 |
ENSRNOT00000016260
|
Dennd4a
|
DENN domain containing 4A |
| chr10_+_59259955 | 0.02 |
ENSRNOT00000021816
|
Ankfy1
|
ankyrin repeat and FYVE domain containing 1 |
| chrX_+_51286737 | 0.02 |
ENSRNOT00000035692
|
Dmd
|
dystrophin |
| chr10_-_72417070 | 0.02 |
ENSRNOT00000036814
|
Usp32
|
ubiquitin specific peptidase 32 |
| chr17_+_15762030 | 0.02 |
ENSRNOT00000089310
|
Fgd3
|
FYVE, RhoGEF and PH domain containing 3 |
| chr4_-_129515435 | 0.02 |
ENSRNOT00000039353
|
Eogt
|
EGF domain specific O-linked N-acetylglucosamine transferase |
| chr2_+_95008477 | 0.02 |
ENSRNOT00000015327
|
Tpd52
|
tumor protein D52 |
| chr4_+_147756553 | 0.02 |
ENSRNOT00000086549
ENSRNOT00000014733 |
Ift122
|
intraflagellar transport 122 |
| chr1_+_274688580 | 0.02 |
ENSRNOT00000020580
|
Shoc2
|
SHOC2 leucine-rich repeat scaffold protein |
| chr5_-_7941822 | 0.02 |
ENSRNOT00000079917
|
Prex2
|
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chr14_+_108143869 | 0.01 |
ENSRNOT00000090003
|
Usp34
|
ubiquitin specific peptidase 34 |
| chr11_+_87522971 | 0.01 |
ENSRNOT00000043545
|
Smpd4
|
sphingomyelin phosphodiesterase 4 |
| chr19_+_49637016 | 0.01 |
ENSRNOT00000016880
|
Bco1
|
beta-carotene oxygenase 1 |
| chr2_+_257626383 | 0.01 |
ENSRNOT00000080993
|
Zzz3
|
zinc finger, ZZ-type containing 3 |
| chr10_-_57291146 | 0.01 |
ENSRNOT00000005653
|
Spag7
|
sperm associated antigen 7 |
| chr8_-_87245951 | 0.01 |
ENSRNOT00000015299
|
Tmem30a
|
transmembrane protein 30A |
| chr15_-_88670349 | 0.01 |
ENSRNOT00000012881
|
Rnf219
|
ring finger protein 219 |
| chr19_-_11669578 | 0.01 |
ENSRNOT00000026373
|
Gnao1
|
G protein subunit alpha o1 |
| chrX_+_84064427 | 0.01 |
ENSRNOT00000046364
|
Zfp711
|
zinc finger protein 711 |
| chr1_-_141655894 | 0.01 |
ENSRNOT00000082591
|
Ap3s2
|
adaptor-related protein complex 3, sigma 2 subunit |
| chr5_+_49311030 | 0.01 |
ENSRNOT00000010850
|
Cnr1
|
cannabinoid receptor 1 |
| chr12_+_47590154 | 0.01 |
ENSRNOT00000045946
|
Git2
|
GIT ArfGAP 2 |
| chr2_-_198852161 | 0.01 |
ENSRNOT00000028815
|
Polr3c
|
RNA polymerase III subunit C |
| chr7_+_141642777 | 0.01 |
ENSRNOT00000079811
|
AABR07058884.2
|
|
| chr6_-_114476723 | 0.01 |
ENSRNOT00000005162
|
Dio2
|
deiodinase, iodothyronine, type II |
| chr14_-_21127868 | 0.01 |
ENSRNOT00000020608
ENSRNOT00000092172 |
Rufy3
|
RUN and FYVE domain containing 3 |
| chr7_+_142877086 | 0.01 |
ENSRNOT00000088730
|
Grasp
|
general receptor for phosphoinositides 1 associated scaffold protein |
| chrX_+_54266687 | 0.01 |
ENSRNOT00000058309
|
LOC100363193
|
LRRGT00076-like |
| chr18_+_55505993 | 0.01 |
ENSRNOT00000043736
|
RGD1309362
|
similar to interferon-inducible GTPase |
| chr4_-_145948996 | 0.01 |
ENSRNOT00000043476
|
Atp2b2
|
ATPase plasma membrane Ca2+ transporting 2 |
| chr3_+_8430829 | 0.01 |
ENSRNOT00000090440
ENSRNOT00000060969 |
Cercam
|
cerebral endothelial cell adhesion molecule |
| chr1_-_53802658 | 0.01 |
ENSRNOT00000032667
|
Afdn
|
afadin, adherens junction formation factor |
| chr18_+_17403407 | 0.01 |
ENSRNOT00000045150
|
RGD1562608
|
similar to KIAA1328 protein |
| chr3_-_121488540 | 0.01 |
ENSRNOT00000023845
|
Zc3h8
|
zinc finger CCCH type containing 8 |
| chr16_+_8737974 | 0.01 |
ENSRNOT00000064255
|
Ercc6
|
ERCC excision repair 6, chromatin remodeling factor |
| chr1_+_73837944 | 0.01 |
ENSRNOT00000036413
|
Lair1
|
leukocyte-associated immunoglobulin-like receptor 1 |
| chrX_+_44907521 | 0.01 |
ENSRNOT00000004901
|
Tbl1x
|
transducin (beta)-like 1 X-linked |
| chr7_-_11699436 | 0.01 |
ENSRNOT00000049221
ENSRNOT00000066037 |
Tmprss9
|
transmembrane protease, serine 9 |
| chr10_+_36589181 | 0.01 |
ENSRNOT00000045596
|
Zfp354a
|
zinc finger protein 354A |
| chr5_-_33664435 | 0.01 |
ENSRNOT00000009047
|
Wwp1
|
WW domain containing E3 ubiquitin protein ligase 1 |
| chrX_-_13279082 | 0.01 |
ENSRNOT00000051898
ENSRNOT00000060857 |
Tspan7
|
tetraspanin 7 |
| chr3_-_90751055 | 0.01 |
ENSRNOT00000040741
|
LOC499843
|
LRRGT00091 |
| chr20_-_28263037 | 0.01 |
ENSRNOT00000030348
|
Edar
|
ectodysplasin-A receptor |
| chr8_+_59900651 | 0.01 |
ENSRNOT00000020410
|
Tmem266
|
transmembrane protein 266 |
| chrX_-_62690806 | 0.01 |
ENSRNOT00000018147
|
Pola1
|
DNA polymerase alpha 1, catalytic subunit |
| chr15_-_33752665 | 0.01 |
ENSRNOT00000034102
|
Zfhx2
|
zinc finger homeobox 2 |
| chr10_-_89130339 | 0.01 |
ENSRNOT00000027640
|
Ezh1
|
enhancer of zeste 1 polycomb repressive complex 2 subunit |
| chr19_+_37576075 | 0.01 |
ENSRNOT00000089022
|
Fam65a
|
family with sequence similarity 65, member A |
| chr10_-_4910305 | 0.01 |
ENSRNOT00000033122
|
Rmi2
|
RecQ mediated genome instability 2 |
| chr8_+_117068582 | 0.01 |
ENSRNOT00000073559
|
Amt
|
aminomethyltransferase |
| chr1_-_102849430 | 0.01 |
ENSRNOT00000086856
|
Saa4
|
serum amyloid A4 |
| chrX_+_135092040 | 0.01 |
ENSRNOT00000048022
|
Utp14a
|
UTP14A small subunit processome component |
| chr1_+_84758233 | 0.01 |
ENSRNOT00000072734
|
AABR07002775.1
|
|
| chr8_-_69121192 | 0.01 |
ENSRNOT00000012404
|
Zwilch
|
zwilch kinetochore protein |
| chr18_-_74059533 | 0.01 |
ENSRNOT00000038767
|
RGD1308601
|
similar to hypothetical protein |
| chr10_-_31419235 | 0.01 |
ENSRNOT00000059496
|
Cyfip2
|
cytoplasmic FMR1 interacting protein 2 |
| chr1_+_264741911 | 0.01 |
ENSRNOT00000019956
|
Sema4g
|
semaphorin 4G |
| chr2_-_22059149 | 0.01 |
ENSRNOT00000031909
|
Fam151b
|
family with sequence similarity 151, member B |
| chr1_-_198900375 | 0.01 |
ENSRNOT00000024969
|
Zfp689
|
zinc finger protein 689 |
| chr13_+_89586283 | 0.01 |
ENSRNOT00000079355
ENSRNOT00000049873 |
Nr1i3
|
nuclear receptor subfamily 1, group I, member 3 |
| chr10_+_61685241 | 0.01 |
ENSRNOT00000092606
|
Mnt
|
MAX network transcriptional repressor |
| chr4_-_163890801 | 0.01 |
ENSRNOT00000081946
|
Ly49si1
|
immunoreceptor Ly49si1 |
| chr12_-_38916237 | 0.01 |
ENSRNOT00000074517
|
Tmem120b
|
transmembrane protein 120B |
| chr10_+_67762995 | 0.01 |
ENSRNOT00000000253
|
Zfp207
|
zinc finger protein 207 |
| chr5_-_88612626 | 0.01 |
ENSRNOT00000089560
|
Tle1
|
transducin like enhancer of split 1 |
| chr2_+_149214265 | 0.01 |
ENSRNOT00000084020
|
Med12l
|
mediator complex subunit 12-like |
| chr7_-_11400805 | 0.01 |
ENSRNOT00000027634
|
Dapk3
|
death-associated protein kinase 3 |
| chr13_+_99005142 | 0.01 |
ENSRNOT00000004293
|
Acbd3
|
acyl-CoA binding domain containing 3 |
| chr1_+_219764001 | 0.01 |
ENSRNOT00000082388
|
Pc
|
pyruvate carboxylase |
| chr12_+_30198822 | 0.01 |
ENSRNOT00000041645
|
Gusb
|
glucuronidase, beta |
| chr2_+_86914989 | 0.01 |
ENSRNOT00000085164
ENSRNOT00000081966 |
LOC100360380
|
zinc finger protein 457-like |
| chr10_-_71849293 | 0.01 |
ENSRNOT00000003799
|
Lhx1
|
LIM homeobox 1 |
| chr14_+_4125380 | 0.01 |
ENSRNOT00000002882
|
Zfp644
|
zinc finger protein 644 |
| chr18_-_37245809 | 0.01 |
ENSRNOT00000079585
|
Ppp2r2b
|
protein phosphatase 2, regulatory subunit B, beta |
| chr7_-_80457816 | 0.01 |
ENSRNOT00000039430
|
AABR07057617.1
|
|
| chr10_+_53621375 | 0.01 |
ENSRNOT00000004147
|
Myh3
|
myosin heavy chain 3 |
| chr5_+_64892802 | 0.01 |
ENSRNOT00000059869
|
Rnf20
|
ring finger protein 20 |
| chr8_+_61671513 | 0.00 |
ENSRNOT00000091128
|
Ptpn9
|
protein tyrosine phosphatase, non-receptor type 9 |
| chr14_+_48726045 | 0.00 |
ENSRNOT00000086646
|
Arap2
|
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr19_+_17115412 | 0.00 |
ENSRNOT00000015876
|
Rpgrip1l
|
Rpgrip1-like |
| chr16_+_2278701 | 0.00 |
ENSRNOT00000016233
|
Dennd6a
|
DENN domain containing 6A |
| chrX_+_115351114 | 0.00 |
ENSRNOT00000047553
|
Hmg1l1
|
high-mobility group (nonhistone chromosomal) protein 1-like 1 |
| chr15_+_51303909 | 0.00 |
ENSRNOT00000085237
ENSRNOT00000058663 |
Loxl2
|
lysyl oxidase-like 2 |
| chr1_+_84584596 | 0.00 |
ENSRNOT00000032933
|
LOC100910046
|
zinc finger protein 60-like |
| chr4_-_108008484 | 0.00 |
ENSRNOT00000007971
|
Ctnna2
|
catenin alpha 2 |
| chr10_+_80790168 | 0.00 |
ENSRNOT00000073315
ENSRNOT00000075163 |
Car10
|
carbonic anhydrase 10 |
| chr2_-_190100276 | 0.00 |
ENSRNOT00000015351
|
S100a9
|
S100 calcium binding protein A9 |
| chr3_-_177078594 | 0.00 |
ENSRNOT00000020695
|
Uckl1
|
uridine-cytidine kinase 1-like 1 |
| chr2_-_111793326 | 0.00 |
ENSRNOT00000092660
|
Nlgn1
|
neuroligin 1 |
| chrX_-_74968405 | 0.00 |
ENSRNOT00000035653
|
RGD1561931
|
similar to KIAA2022 protein |
| chr17_-_15429322 | 0.00 |
ENSRNOT00000093381
|
Nol8
|
nucleolar protein 8 |
| chr4_-_119568736 | 0.00 |
ENSRNOT00000041234
|
Aplf
|
aprataxin and PNKP like factor |
| chr2_+_44512895 | 0.00 |
ENSRNOT00000013218
|
Slc38a9
|
solute carrier family 38, member 9 |
| chr17_-_14589154 | 0.00 |
ENSRNOT00000073035
|
Nol8
|
nucleolar protein 8 |
| chr2_-_198706428 | 0.00 |
ENSRNOT00000085006
|
Polr3gl
|
RNA polymerase III subunit G like |
| chr1_+_193844538 | 0.00 |
ENSRNOT00000065311
|
AC123018.1
|
|
| chr17_-_15429051 | 0.00 |
ENSRNOT00000074667
|
Nol8
|
nucleolar protein 8 |
| chr10_-_89700283 | 0.00 |
ENSRNOT00000028213
|
Etv4
|
ets variant 4 |
| chr4_+_96831880 | 0.00 |
ENSRNOT00000068400
|
RSA-14-44
|
RSA-14-44 protein |
| chr10_-_12871965 | 0.00 |
ENSRNOT00000004583
|
Gm8225
|
predicted gene 8225 |
| chr20_+_25990304 | 0.00 |
ENSRNOT00000033980
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3 |
| chr1_-_141504111 | 0.00 |
ENSRNOT00000040164
|
Snrpep2
|
small nuclear ribonucleoprotein polypeptide E pseudogene 2 |
| chr9_+_44681494 | 0.00 |
ENSRNOT00000035785
|
Eif5b
|
eukaryotic translation initiation factor 5B |
| chr1_+_256955652 | 0.00 |
ENSRNOT00000020411
|
Lgi1
|
leucine-rich, glioma inactivated 1 |
| chr17_+_69365453 | 0.00 |
ENSRNOT00000023133
|
Akr1e2
|
aldo-keto reductase family 1, member E2 |
| chr15_-_27798408 | 0.00 |
ENSRNOT00000088981
|
Tep1
|
telomerase associated protein 1 |
| chr2_-_258932200 | 0.00 |
ENSRNOT00000045905
|
Adgrl2
|
adhesion G protein-coupled receptor L2 |
| chr3_+_129599353 | 0.00 |
ENSRNOT00000008734
|
Snap25
|
synaptosomal-associated protein 25 |
Network of associatons between targets according to the STRING database.
First level regulatory network of Onecut2_Onecut3
Gene Ontology Analysis
Gene overrepresentation in biological process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015766 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0097680 | double-strand break repair via classical nonhomologous end joining(GO:0097680) |
| 0.0 | 0.1 | GO:0035426 | extracellular matrix-cell signaling(GO:0035426) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0036371 | protein localization to M-band(GO:0036309) protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.0 | GO:0070662 | negative regulation of mast cell apoptotic process(GO:0033026) mast cell proliferation(GO:0070662) regulation of mast cell proliferation(GO:0070666) positive regulation of mast cell proliferation(GO:0070668) |
| 0.0 | 0.1 | GO:0031394 | positive regulation of prostaglandin biosynthetic process(GO:0031394) positive regulation of unsaturated fatty acid biosynthetic process(GO:2001280) |
| 0.0 | 0.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.0 | GO:0046103 | inosine biosynthetic process(GO:0046103) |
Gene overrepresentation in cellular component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032807 | DNA ligase IV complex(GO:0032807) |
| 0.0 | 0.0 | GO:0031230 | intrinsic component of cell outer membrane(GO:0031230) integral component of cell outer membrane(GO:0045203) |
Gene overrepresentation in molecular function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0015157 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.1 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.1 | GO:0043237 | laminin-1 binding(GO:0043237) |